Figure 3.
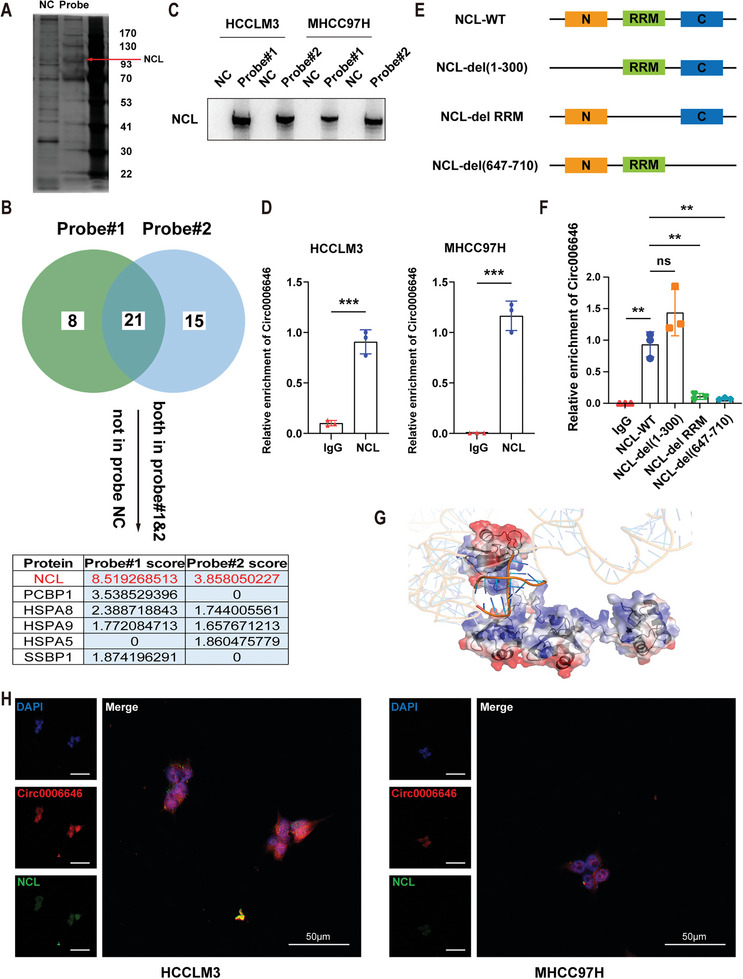
Circ0006646 combined with NCL in HCC cells. A) After cell lysis using RIPA, circ0006646 specific targeting probe and NC probe were used for pulldown assay, followed by protein silver staining. B) Selection strategies for potential binding proteins. After mass spectrometry, peptides which presented in two circ0006646 specific targeting probe groups but not in the NC probe group were considered as candidates for circ0006646 binding proteins. Select the one with the highest score for follow‐up verification. C) NCL enriched by two circ0006646 specific probes was verified. Blots with antibodies recognizing the NCL and β‐Actin were shown. D) The enrichment of circ0006646 in HCCLM and MHCC97H was detected by using NCL antibody and IgG antibody in RIP assay. E) Sketch‐map of three NCL truncated mutations. C: C‐terminal; N: n‐terminal; RRM: RNA recognition motif. F) Four different NCL plasmids (wild type and truncated mutation) were transfected into cells. In RIP assay, NCL antibody and IgG antibody were used to measure the enrichment of circ0006646. G) Graphical representation of a complex formed by the circ0006646 and NCL307‐647. The semi‐transparent blue–white–red surface depicts protein's contact potential (blue: positively charged, red: negatively charged). H) Representative IF images about the binding of circ0006646 to NCL were shown. DAPI staining represented the location of the nucleus. Red: circ0006646, green: NCL. All experiments were performed in at least triplicate samples. Data were presented as the means ± SD. ** p < 0.01. Student's t‐test was used for normally distributed data and Mann–Whitney U test was used for skewed distribution data.
