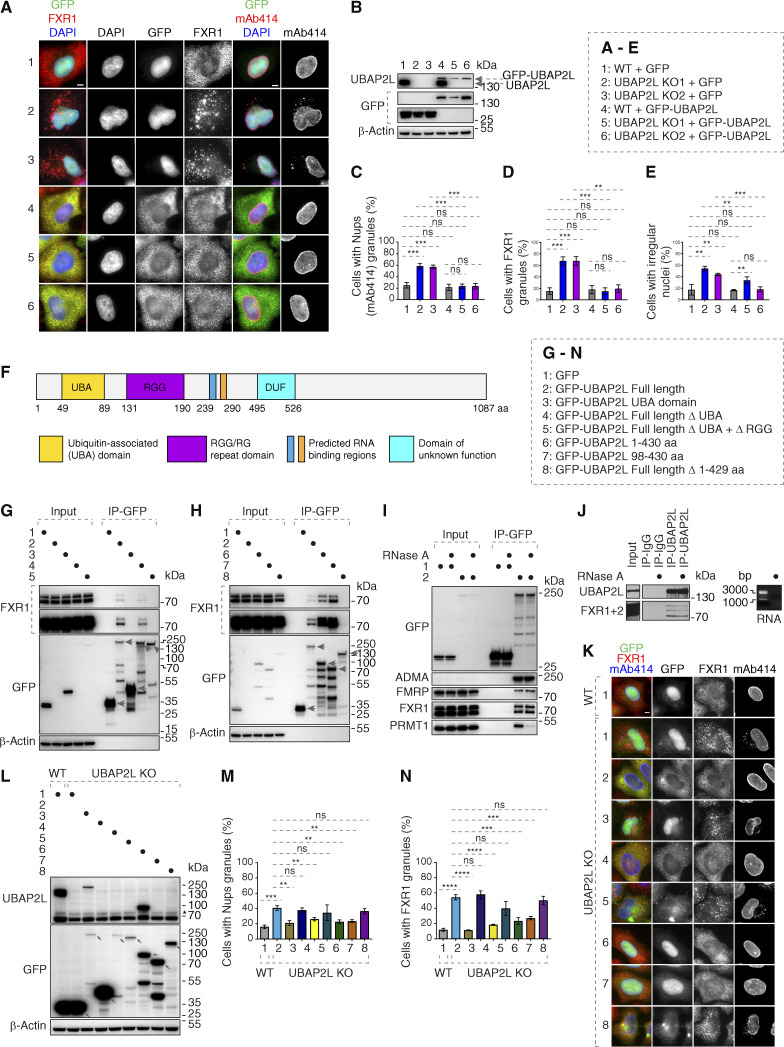
Figure S4.
98–430 aa fragment of UBAP2L protein is required for the function of UBAP2L on Nups and FXR1. (A–E) Representative immunofluorescence images depicting the nuclear shape and localization of FXR1 and Nups (mAb414) in WT and UBAP2L KO HeLa cells expressing GFP alone or GFP-UBAP2L for 60 h and synchronized in interphase by DTBR at 12 h. Nuclei were stained with DAPI. Scale bars, 5 μm. Note that ectopic expression of GFP-UBAP2L but not GFP can rescue the nuclear and localization phenotypes in both UBAP2L KO HeLa cell lines (A). The protein levels of endogenous UBAP2L, GFP, and GFP-UBAP2L of cells shown in A were analyzed by western blot (B). The percentage of cells with the cytoplasmic granules of Nups (mAb414) (C) and of FXR1 (D) and with irregular nuclei (E) shown in A were quantified. At least 200 cells per condition were analyzed (mean ± SD, ns: not significant, **P < 0.01, ***P < 0.001, unpaired two-tailed t test, n = 3 independent experiments). (F) Domain organization of UBAP2L depicting UBA domain, RGG domain, two predicted RNA binding regions, and the domain of unknown function (DUF). (G and H) Lysates of HeLa cells expressing GFP alone or GFP-UBAP2L–dervied constructs (full-length [FL], UBA, ΔUBA, or Δ(UBA+RGG) fragments) for 27 h were immunoprecipitated using agarose GFP-Trap A beads (GFP-IP) and analyzed by western blot (G). Lysates of HeLa cells expressing GFP alone or several GFP-UBAP2L–derived constructs (FL, 1–430 aa, 98–430 aa, or Δ1–429 aa fragments) for 27 h were immunoprecipitated using agarose GFP-Trap A beads (GFP-IP) and analyzed by western blot (H). The arrows indicate the bands corresponding to the expressed GFP proteins while the remaining bands are non-specific. (I and J) Interphase HeLa cells expressing GFP alone or GFP-UBAP2L for 27 h and cell lysates were treated with RNase A, immunoprecipitated using agarose GFP-Trap A beads (GFP-IP), and analyzed by western blot. Note that RNase treatment can abolish interaction with PRMT1 but not with FXRPs (I). IPs from cell lysates of HeLa cells treated with RNase A using UBAP2L antibody or IgG were analyzed by western blot. The efficiency of the RNase treatment was confirmed by imaging of mRNAs by agarose gel electrophoresis and ethidium bromide staining (J). (K–N) Representative immunofluorescence images depicting localization of FXR1 and Nups (mAb414) in WT and UBAP2L KO HeLa cells expressing GFP alone or GFP-UBAP2L-derived fragments (FL, UBA, ΔUBA, Δ(UBA+RGG), 1–430 aa, 98–430 aa, or Δ1–429 aa) for 60 h and synchronized in interphase by DTBR at 12 h. Scale bar, 5 μm (K). Note that the UBAP2L 98–430 aa protein fragment containing the RGG domain is required for the function of UBAP2L on Nups and FXR1. The protein levels of endogenous UBAP2L, GFP, and GFP-UBAP2L-derived versions (FL, UBA, ΔUBA, Δ(UBA+RGG), 1–430 aa, 98–430 aa, or Δ1–429 aa) of cells shown in K were analyzed by western blot. The arrows indicate the bands corresponding to the expressed GFP proteins while the remaining faster migrating bands are either non-specific or degradation products (L). The percentage of cells with the cytoplasmic granules of Nups (mAb414) (M) and of FXR1 (N) shown in K were quantified. At least 200 cells per condition were analyzed (mean ± SD, ns: not significant, **P < 0.01, ***P < 0.001, ****P < 0.0001, unpaired two-tailed t test, n = 3 independent experiments). Source data are available for this figure: SourceData FS4.