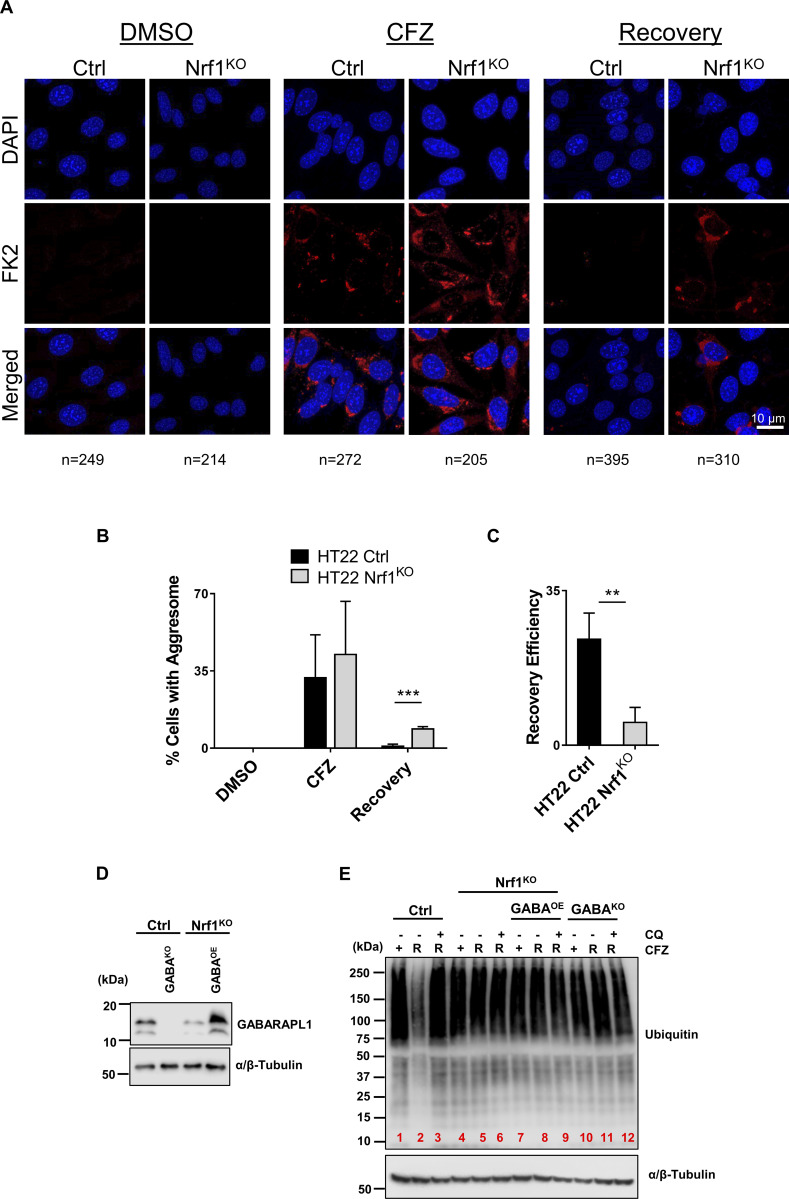
Figure 6.
Nrf1 is required to clear aggresomes that are triggered by proteasome inhibition. (A) HT22 wild-type (control; ctrl) and Nrf1KO cells were treated with 50 nM CFZ for 20 h, then were washed and incubated with fresh media for another 20 h (Recovery period). Aggresomes were detected by confocal microscopy using FK2 stain. Scale bar represents 10 μm. (B) Percentage of cells with aggresomes under each condition (DMSO, CFZ and recovery) for both ctrl and Nrf1KO cells are plotted, based on confocal microscopy analysis. The number of cells analyzed in each sample is noted underneath the images in A. (C) Recovery efficiency of ctrl and Nrf1KO cells was calculated by dividing CFZ values by recovery values of ctrl and Nrf1KO cells in B. (D) Western blot analysis of GABARAPL1 overexpression in Nrf1KO cells. α/β-Tubulin was used as the loading control. (E) HT22 control (EV), GABAKO (GABARAPL1 knockout), Nrf1KO, Nrf1KO GABAOE (GABARAPL1 overexpression) cells were treated with 50 nM CFZ for 20 h, then washed, and either treated with 60 μM CQ or fresh complete media for a 20 h recovery (R). The samples were analyzed by immunoblotting with anti-ubiquitin antibody. α/β-Tubulin was used as the loading control. Three biological replicates were used for confocal microscopy analysis. P values were calculated by Student’s t test. **<0.05, ***<0.005. Source data are available for this figure: SourceData F6.