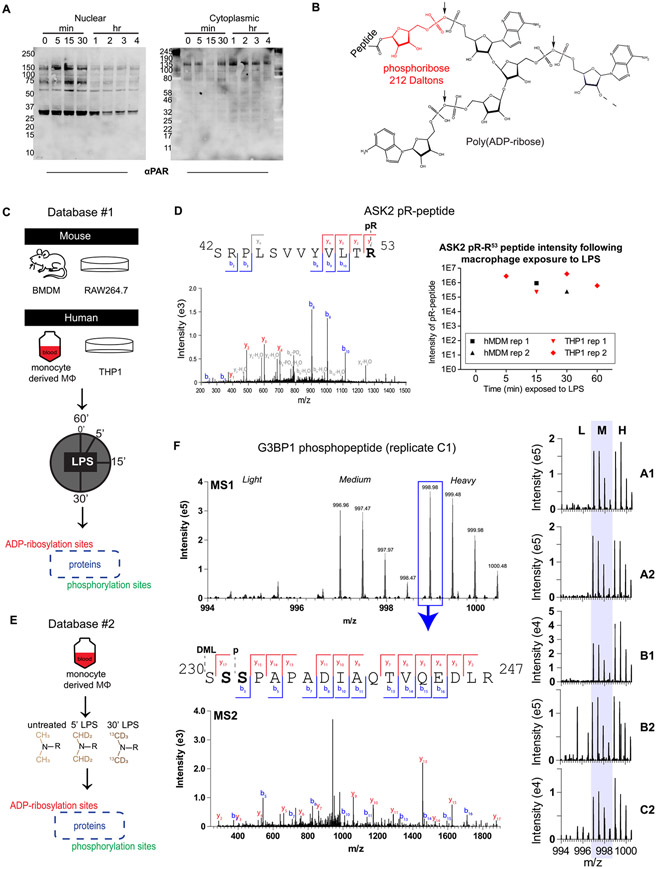
Figure 1.
Assessment of the ADP-ribosylated and phosphorylated proteomes in LPS-stimulated macrophages and generation of Database #1 and Database #2. Nuclear-cytoplasmic fractionation of LPS-stimulated Raw264.7 cells (A) reveals changes in global PARylation patterns during LPS stimulation (100 ng/mL; LPS from S. minnesota). To detect site-specific changes in ADP-ribosylation, snake venom phosphodiesterase was used to cleave the monomer or polymer down to its phosphoribose (pR) attachment site (B) following 100 ng/mL LPS stimulation of human or mouse macrophages for various amounts of time (C). Site identification was achieved through localization of the 212 Da pR group, and PTM dynamics was measured from extracted ion chromatograms (XICs) of modified peptides (example from the ASK2 protein shown in (D), z = 2, m/z = 801.42, source = hMDM rep 1). Primary human monocyte-derived macrophages (hMDMs) were further characterized using dimethyl labeling (DML) to achieve robust relative quantification (E). DML-enabled quantification of an example peptide from replicate C1 of Database #2 is shown in (F); the base peak from the heavy labeled form was selected for MS/MS, and fragmentation reveals that this isolated peak is either a combination of S231 and S232, or a single form that cannot be distinguished. The summed MS1 spectra from other replicates are shown on the right, indicating that this peptide is identified exclusively or primarily in the medium and heavy (LPS-treated) forms. A/B/C indicates a biological replicate (unique human donor), and 1/2 indicates technical replicate (unique derivation of macrophages from blood monocytes). BMDM, bone-marrow-derived macrophages from mice; hMDM, human monocyte-derived macrophages from healthy human donor blood.