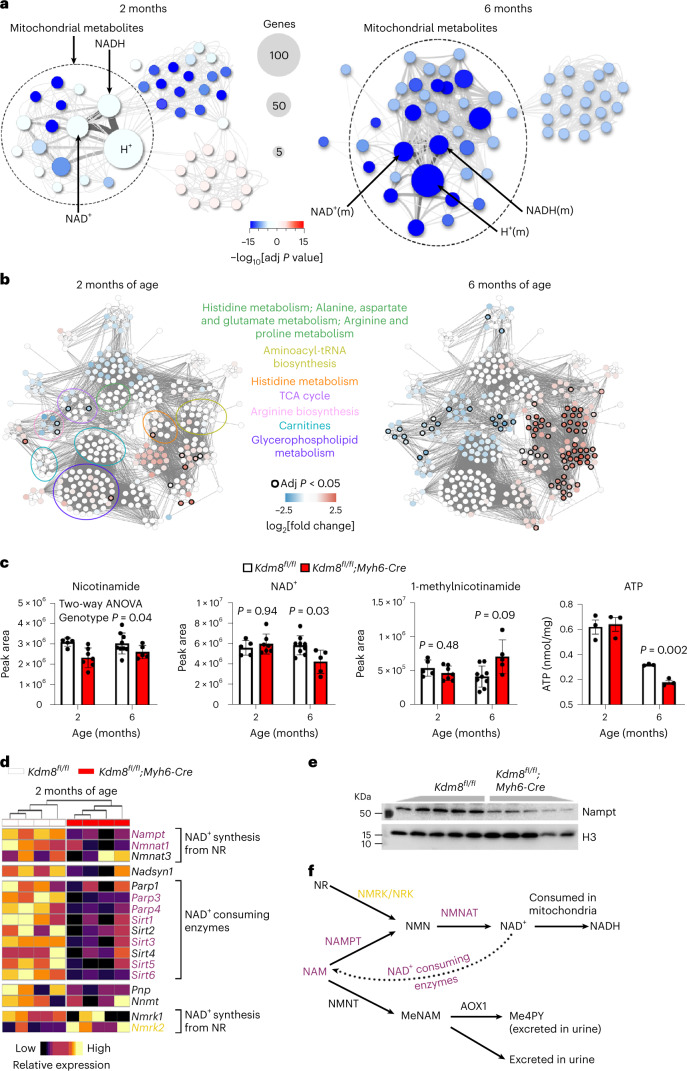
Fig. 3. NAD+ synthesis metabolome is progressively altered at early stages of DCM.
a, Reporter metabolite analysis using differentially expressed genes in ventricles of Kdm8fl/fl control and Kdm8fl/fl;Myh6-Cre mutant mice at 2 and 6 months of age. The left panel illustrates all 46 reporter metabolites detected in 2-month-old hearts. The right panel illustrates the top 55 out of 718 reporter metabolites. Upregulated metabolites are in red, and downregulated metabolites are in blue. Data were analyzed by piano using distinct-directional P values. n = 2 mice per group at 2 months, and 3 mice per group at 6 months. b, Metabolomic networks from liquid chromatography–mass spectrometry (LC–MS) analysis of ventricles of control and mutant hearts at 2 and 6 months of age. Significantly altered metabolites are circled in black. Upregulated metabolites are in red, and downregulated metabolites are in blue. Each time point was independently analyzed by two-tailed Student’s t-test. P values were adjusted for false discovery by the Benjamini–Hochberg method. n = 5 controls and 7 mutants at 2 months; n = 9 controls and 5 mutants at 6 months. c, Quantification of NAM, NAD+ and 1-methyl-NAM by LC–MS, and ATP by fluorometric assay. Data are mean ± s.d. Each time point was analyzed by an independent two-tailed Student’s t-test, and two-way ANOVA with genotype, time point, and genotype–time point interaction as main effects. P values were adjusted for FDR by the Benjamini–Hochberg method. n = 5 controls and 7 mutants at 2 months; n = 9 controls and 5 mutants at 6 months for LC–MS. n = 3 hearts per group for ATP fluorometric assay. d, Heatmap of qPCR of genes encoding enzymes in the NAD+ pathway in 2-month-old ventricles. n = 4 hearts per group for the top 15 genes. n = 5 for Nmrk1 and Nmrk2. Significantly downregulated genes are in purple, and significantly upregulated genes are in yellow. e, Western blot of Nampt in 6-month-old ventricles. n = 5 hearts per group. The experiment was repeated twice with similar results. f, NAD+ biosynthetic pathways highlighting significantly downregulated and upregulated genes in Kdm8 mutant hearts. NAM, nicotinamide; NMN, nicotinamide mononucleotide; NR, nicotinamide riboside; MeNAM, methylnicotinamide; NAD+, nicotinamide adenine dinucleotide; Me4PY, N-methyl-4-pyridone-5-carboxamide.