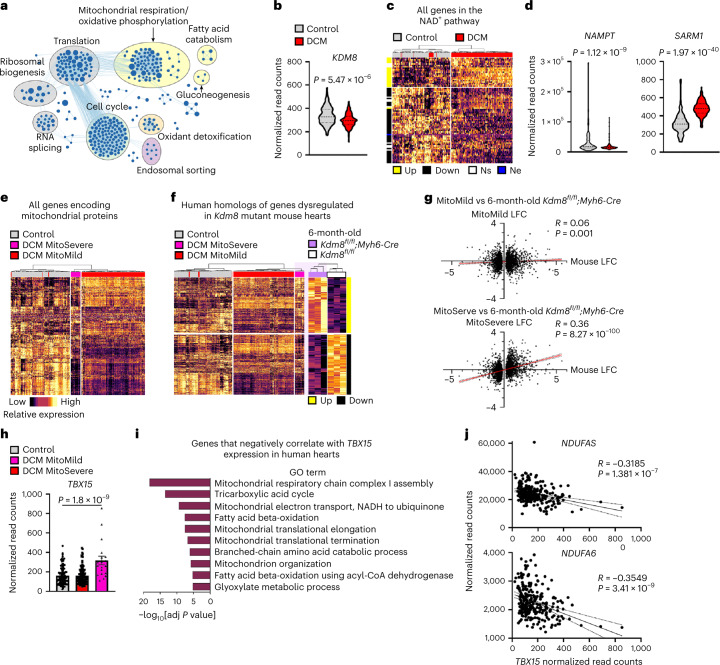
Fig. 6. TBX15 upregulation tracks with stronger downregulation of mitochondrial metabolism regulatory genes in human hearts affected by DCM.
a, GSEA comparing 149 DCM-affected and 113 control human hearts. Each circle represents a gene set, and blue circles represent downregulated gene sets. All gene sets with Q-value < 0.01 are shown. b, Normalized expression of KDM8 in control and DCM-affected hearts. Thick dotted lines denote the median, and thin dashed lines denote the quartiles. Data were analyzed using DESeq2. c, Unsupervised hierarchal clustering of genes in the NAD+ pathway. Up, significantly upregulated; Down, significantly downregulated; Ns, not significant; Ne, not expressed. d, Normalized expression of NAMPT and SARM1 in control and DCM-affected hearts. Thick dotted lines denote the median, and thin dashed lines denote the quartiles. Data were analyzed using DESeq2. e, Unsupervised hierarchal clustering of all genes that encode mitochondrial proteins (obtained from GO term Cellular Component). Three clusters are identified. MitoSevere clusters DCM samples with the lowest expression of mitochondrial genes. f, Unsupervised hierarchal clustering of genes dysregulated in 6-month-old mouse hearts (end-stage DCM) shown to the right, and their human homologs shown to the left. The MitoSevere expression profile closely resembles the 6-month-old mutant hearts (light purple background). g, Pearson correlation between the log2[fold change] (LFC) of dysregulated genes in 6-month-old mutant hearts and of their homologs in the MitoMild (top) or MitoSevere (bottom) samples. Data were analyzed using Pearson correlation two-tailed test. h, Normalized expression of TBX15 in control, MitoMild and MitoSevere hearts. Data are mean ± s.e.m. One-way ANOVA with Tukey’s multiple comparison. n = 113 Control, 130 MitoMild, and 19 MitoSevere transcriptomes. i, GO term enrichment amongst genes whose expression negatively corelated with TBX15 expression across all transcriptomes. Correlation was analyzed using Pearson correlation (two-tailed) with Benjamini and Hochberg multiple comparison correction. n = 262 transcriptomes. GO enrichment was analyzed using DAVID v6.8 with the Benjamini correction. j, Pearson correlation of the expression of TBX15 with NDUFA5 and NDUFA6 across all 262 transcriptomes. The dotted line indicates the 95% confidence interval. Data were analyzed using Pearson correlation (two-tailed) test.