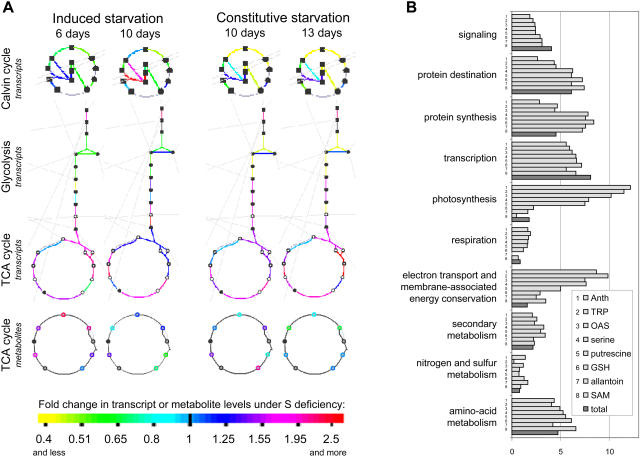
Figure 5.
Integrated analysis of transcriptome and metabolome. A, Mapping of the transcript and metabolite ratios (sulfur-starved plants to control plants) on metabolic pathways from the AraCyc database, using an Omics Viewer tool. Those pathways are depicted, which show the strongest and the most consistent changes on transcript level, and thus are considered to be mostly regulated transcriptionally. B, Functional categorization of the genes, correlating significantly (P < 0.05) to the sulfur-responding metabolites anthocyanins (Anth), tryptophan (TRP), OAS, Ser, putrescine, glutathione (GSH), allantoin, and SAM, depicted as percentage of genes from the corresponding functional category among the whole set of genes, significantly correlating to the corresponding metabolite. These results are put together in comparison to the percent representation of the corresponding functional category in the total set of annotated genes (dark columns).