Figure 4. Mitotic crossovers in Blm mutants.
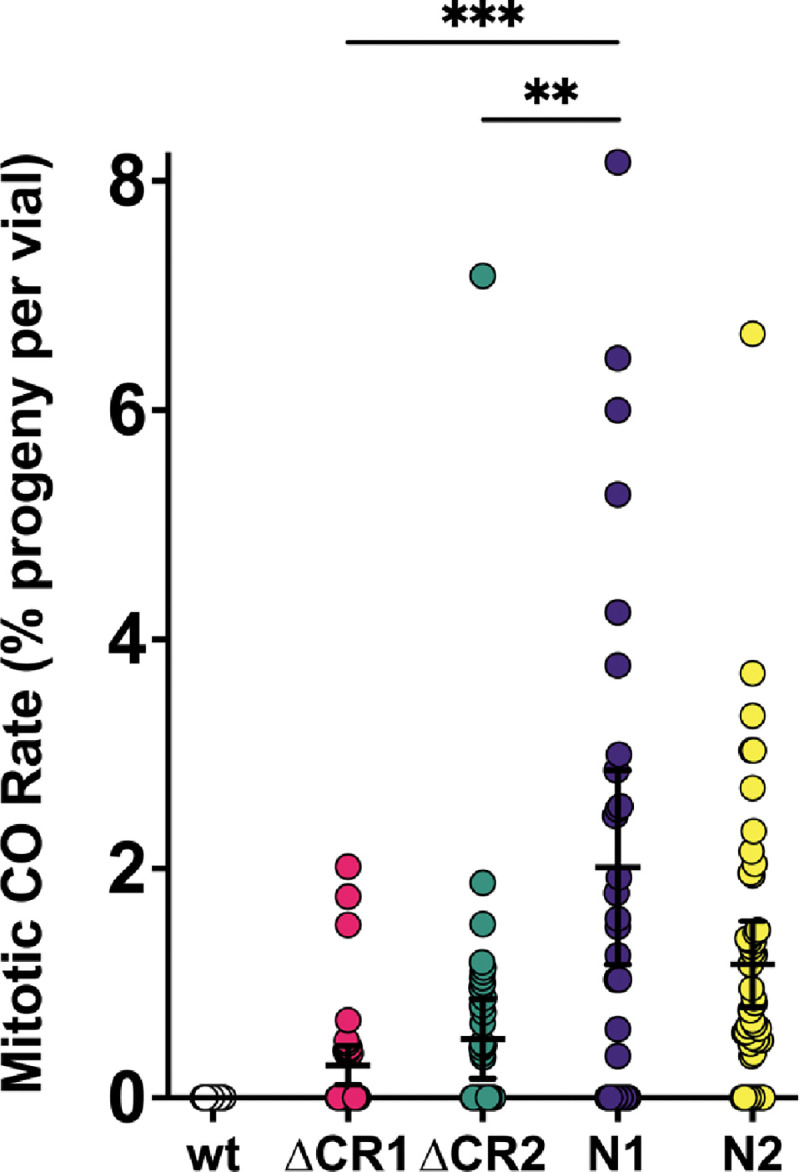
Single males with the Blm alleles indicated on the X-axis over a the BlmN1 null allele (or BlmD2 for BlmN1) were crossed to homozygous net dppho dpy b pr cn recessive phenotypic marker virgin females, with each vials serving as a biological replicate. Progeny were then scored for mitotic crossovers occurring in the parental male’s germline, indicated by mixed presence and/or absence of recessive phenotypes. To obtain the mitotic crossover rate per vial, the number of mitotic crossover progeny was divided by the total number of progeny in that vial. Rates for each vial were then pooled to obtain a mean mitotic crossover rate for each genotype. Crossovers are extremely rare in wild-type males (3), so these are excluded from statistical analyses. While both BlmΔCR1 (ΔCR1) and BlmΔCR2 (ΔCR2) have mitotic crossovers, the rates in both mutants are significantly less than that of the BlmN1 null mutants (N1). ***p<0.001 and **p < 0.01 by ANOVA with Tukey’s Post Hoc. Compared to the separation-of-function BlmN2 (N2; n = 7390) mutant, ΔCR1 mutants had significantly fewer mitotic crossovers (p < 0.05 by ANOVA with Tukey’s Post Hoc), but ΔCR2 was not significantly different. n = wt: 37 vials, 7091 progeny; ΔCR1: 54 vials, 9284 progeny; ΔCR2: 44 vials, 7174 progeny; N1: 88 vials, 9368 progeny; N2: 54 vials, 7390 progeny.
