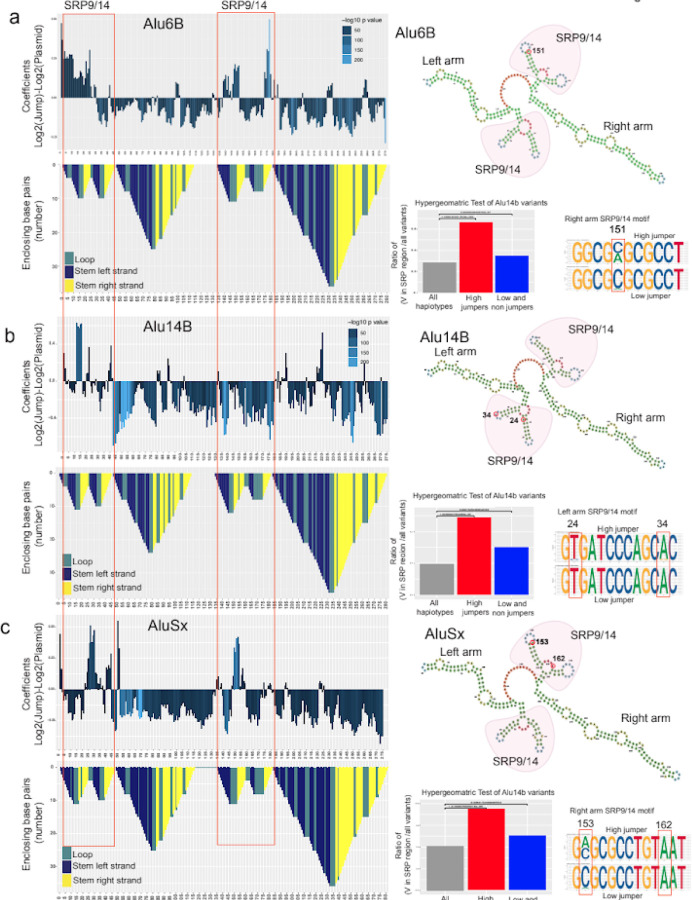
Figure 4: Mutations that affect jumping are associated with SRP binding domains.
a-c, In the top panel 5bp-sliding window analyses of Alu6B (a), Alu14b (b), and AluSx (c), are shown along with the Alu-RNA folding structure in the panels below. The folding structure is reverse mountain coded with different colors for each secondary structure: stem loop-unhybridized in teal, hybridized left strand of the stem in dark blue and hybridized right arm of the stem in yellow. The upper right panel show predicted RNA secondary structures with the left SRP and right arm SRP binding folds highlighted in red. The right lower panels show hypergeometric analyses with the enrichment of variants of dominant jumping effect in SRP binding regions (red bar) versus the enrichment of variants of negative jumping effect in SRP binding regions (blue bar). Next to these, sequence logos show DNA nucleotide composition observed in high jumper (top) and low jumper (bottom) haplotypes at select positions.