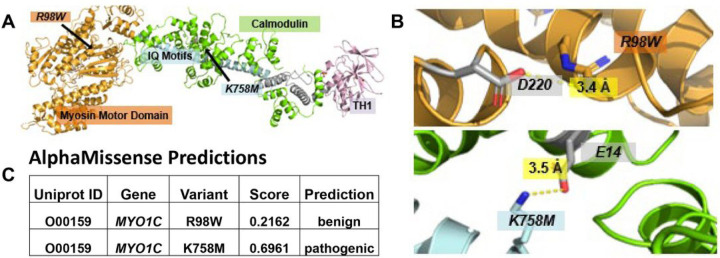
Figure 2.
Crystallized structures of MYO1C (PDB: 4byf, 4r8g). Structural modeling was performed in molecular viewer PyMol using the super function aligning shared regions within both partial structures. In both 4byf and 4r8g structures, calmodulin, a common regulator of myosin activity, is shown to bind to the IQ motif. (A) Complete MYO1C protein structure modeled next to calmodulin. (B) Close-up view of molecular residue interactions at both mutation sites. At both residue sites (K758, R98), there are ion-ion interactions stabilizing intradomain and ligand interactions: R98 binds to nearby D220 within the Myosin Motor Domain and K758 binds to E14 on a calmodulin molecule. Mutations of these charged residues to non-charged amino acids could ablate these ionic interactions, weakening protein structure and function. (C) AlphaMissense analyzes the effects of missense variants on protein tertiary structure. It predicts K758M to be a pathogenic mutation with a score of 0.69