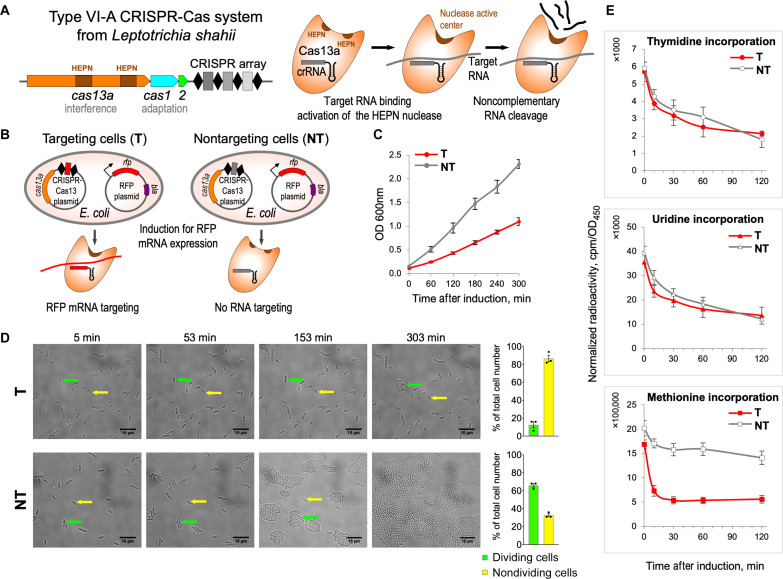
Fig. 1. Cell growth is retarded and translation is inhibited in E. coli cells harboring LshCas13a activated by a nonessential target RNA.
(A) Schematic of the L. shahii CRISPR-Cas13a locus (left) encoding the Cas13a protein with two HEPN domains involved in interference, the Cas1 and Cas2 adaptation proteins, and a CRISPR array. Cas13a collateral RNase activity (right): Target RNA binding to cognate crRNA activates the HEPN nuclease leading to cleavage of noncomplementary RNA. (B) RFP mRNA targeting by LshCas13a in E. coli. Cells contain the CRISPR-Cas13 plasmid that encodes LshCas13a and crRNA and the RFP plasmid that directs inducible expression of RFP. In targeting cells (T), CRISPR array contains a spacer (shown in red) matching a site in RFP mRNA; in nontargeting control cells (NT), the CRISPR array spacer (shown in gray) does not match any cellular RNA. (C) Targeting of nonessential RFP mRNA by LshCas13a leads to growth retardation in targeting (T, red) cells; no such effect is observed in nontargeting (NT, gray) control. OD600 (optical density at 600 nm) values represent means ± SEM of three biological replicates. (D) Time-lapse microscopy showing dividing (green arrows) and nondividing (yellow arrows) cells. Bar graph shows mean percentages (± SEM, n = 3) of dividing and nondividing cells from the total number of cells counted for targeting (top graph) and nontargeting (bottom graph) samples. At least 100 cells were analyzed for each experiment. (E) Time course showing incorporation of radiolabeled precursors: [3H]-thymidine, [3H]-uridine, or [35S]-methionine by pulse labeling in targeting and nontargeting E. coli cells. Radioactivity at each time point is normalized to OD450 of the samples taken after RFP induction. Means ± SEM, n = 3 are shown.