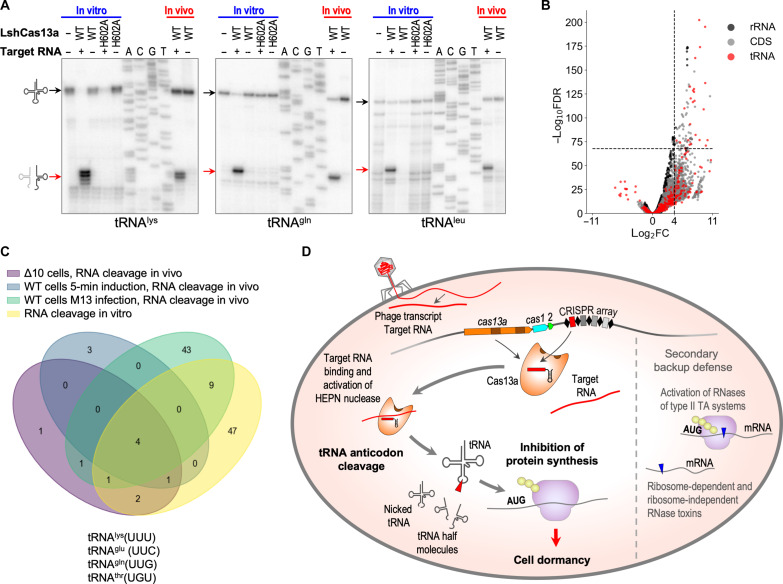
Fig. 5. tRNA cleavage is the prime mechanism of CRISPR-Cas immunity in type VI-A system of L. shahii.
(A) Target-activated Cas13a cuts tRNA at the anticodon loop in vitro. Bulk tRNA was used as a substrate for cleavage. Cleavage products revealed by primer extension following in vitro cleavage by target-activated LshCas13a match those observed in vivo. As expected, the H602A mutation of the catalytic residue in the HEPN1 domain abolishes LshCas13a cleavage activity. Black arrows show full-size tRNAs, and red arrows indicate cleavage products. (B) Volcano plot depicting the results of the comparison of 5′-end counts per nucleotide position of each strand between targeting and nontargeting samples in in vitro cleavage experiments with isolated total E. coli RNA. The analysis was performed in the same way as for Fig. 2H. (C) tRNAlys, tRNAglu, tRNAgln, and tRNAthr are the primary substrates for target-activated Cas13a cleavage in vivo and in vitro. Venn diagram shows the overlaps between the sets of RNA cleavage sites detected by RNA-seq from the four experimental systems used in this work: targeting of plasmid encoded RFP mRNA in wild-type (5 min after induction) and Δ10 (60 min after induction) E. coli cells; phage RNA targeting in M13-infected cells; and RNA cleavage by LshCas13a in vitro. For each dataset, 5′ transcript ends counts with log2FC value between targeting and nontargeting samples > 4 were selected from top 100 hits sorted by adjusted P values in ascending order. Resulting sets of RNA cleavage sites were used to build Venn diagram. (D) Model of defense provided by CRISPR-Cas13a system of L. shahii.