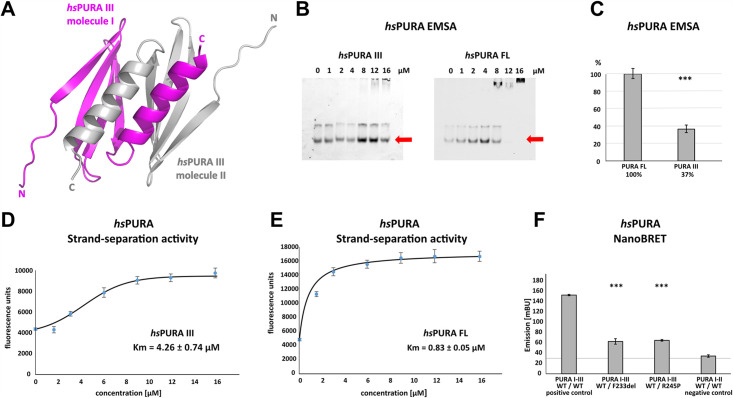
Figure 3. Crystal structure of C-terminal PUR domain from hsPURA and effects of mutations on its function.
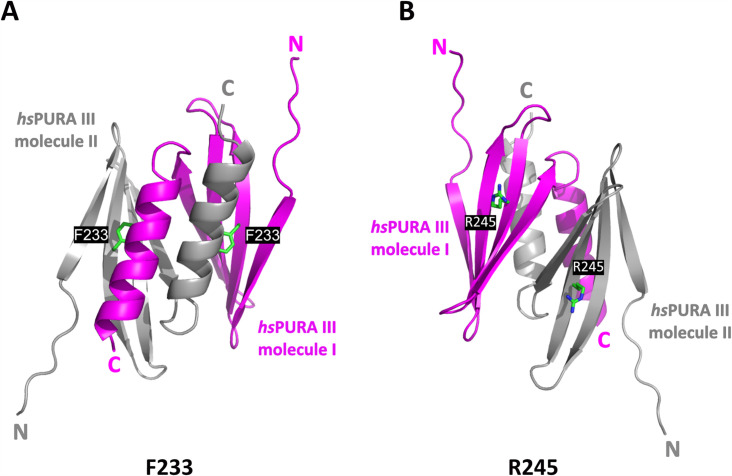
(A) Crystal structure of hsPURA III at 1.7 Å resolution (PDB ID: 8CHW). Two hsPURA III molecules (magenta and gray, respectively) form a homodimer. Like other PUR domains (e.g. hsPURA I–II), hsPURA repeat III consists of four β-sheets and one α-helix. (B) In electrophoretic mobility shift assays (EMSAs) the interaction of hsPURA FL and hsPURA III with a 24-mer (CGG)8 RNA fluorescently labeled with Cy5 fluorophore at 5′-end was observed. The amount of the RNA was kept constant at 8 nM while the protein concentration increased from 0 to 16 μM. The unbound RNA used for quantification (see E) is indicated with red arrows. (C) Apparent affinities derived from EMSAs (C) indicate that the C-terminal PUR domain is also able to interact with the nucleic acids. Pairwise t-tests of the hsPURA fragments showed significantly lower RNA binding compared to the full-length protein (hsPURA III: p = 9.6E−4). Three replicates were measured for each experiment and protein variant, and the standard deviations have been calculated and shown as bars. (D, E) Strand-separation activity of hsPURA. The graphs show the averaged values of three independent experiments as dots with standard deviations as error bars. (D) The hsPURA III fragment shows sigmoidal increase of the ssDNA concentration measured as a fluorescent signal. For the quantification of the sigmoidal curves, we utilized Boltzmann function implemented in the Origin software. Calculated x0, which corresponds to the Km in this assay yields 4.26 ± 0.74 µM. (E) Strand-separation activity of full-length hsPURA was calculated with Michaelis–Menten kinetic, yielding an average Km value of 0.83 ± 0.05 µM, respectively. For both hsPURA samples at least three independent measurements have been performed. (F) NanoBRET experiments in HEK293 cells with different hsPURA fragment-expressing constructs. Milli-BRET units (mBU) were measured for dimerization of hsPURA I–III with hsPURA I–III, hsPURA I–III F233del, and hsPURA I–III R245P. Pairwise t-tests of the mutant hsPURA I–III versions F233del and R245P yielded significantly lower signals compared to the wild-type protein (hsPURA I–III F233del: p = 3.3E−4; hsPURA I–III R245P: p = 1.5E−7), indicating impaired interactions between the proteins. Black horizontal line shows reference of mBU obtained for hsPURA I–II as negative control. Of note, since the BRET signal is the ratio of donor and acceptor signal, it does not require normalization for expression levels. Asterisks in (C) and (F) indicate significance level: *** for p ≤ 0.001.