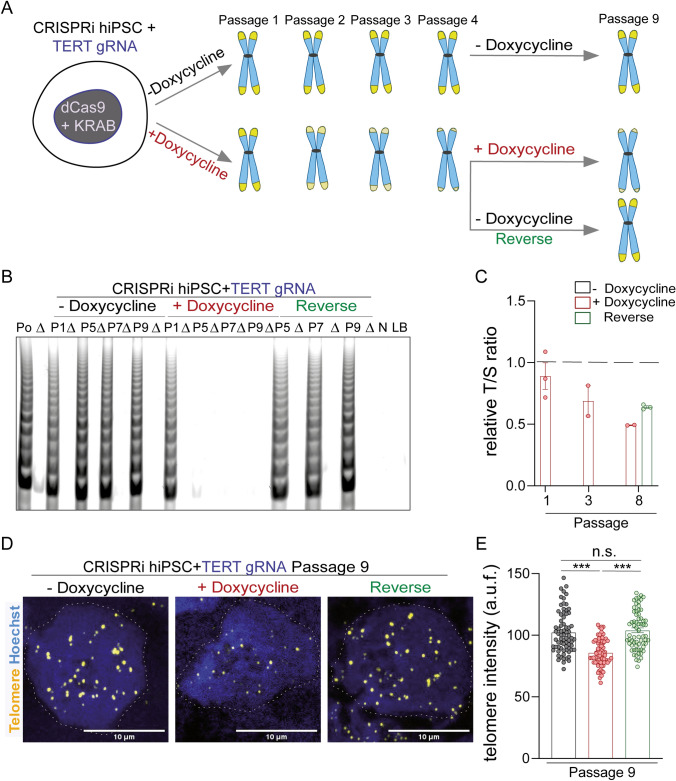
Fig. 1.
Modulation of telomerase and telomere lengths in hiPSCs using CRISPRi. A Schematic representation of the telomerase modulation in the CRISPRi TERT hiPSCs using doxycycline induction and reversal of telomerase modulation by withdrawal of doxycycline. The replicative shortening (and recovery in reverse experiment strategy) of telomere length is expected to occur gradually through subsequent passaging of the cells. B Telomerase activity in cell lysates of CRISPRi TERT hiPSCs under doxycycline treatment was negligible from passage 5 (+ Doxycycline), which could be rescued back to untreated (− Doxycycline) control levels after removal of doxycycline treatment. (n = 200,000 cells per group), Po indicates cell lysate of TRAP positive sample (hiPSCs); ∆ indicates heat inactivated cell lysate; P indicates passage of the hiPSCs; N indicates cell lysate of TRAP negative sample (HUVEC); LB indicates lysis buffer. C The fold change in telomere lengths of CRISPRi TERT hiPSCs as measured by qPCR. Data shows reduced telomere lengths after CRISPRi induction (+ Doxycycline, red bar) and further rescue upon removal of doxycycline induction (Reverse, green bar). The dotted black line indicates telomere length of the control (− Doxycycline) CRISPRi TERT iPSCs. (n = 2–3 sample per group). D–E Representative images and quantification performed for CRISPRi TERT hiPSCs at passage 9 using TEL-qFISH. Shortening of telomere lengths is observed upon doxycycline-mediated CRISPRi induction (+ Doxycycline, red bar) which is reversed after removal of doxycycline (Reverse, green bar) (n ≥ 66 nuclei per group). Scale bar indicates 10 µm; The dotted grey line indicates the nuclei region used for qFISH analysis. hiPSCs = human induced pluripotent stem cells; *** p < 0.001; One-way ANOVA, Kruskal–Wallis test with Dunn’s multiple comparison test