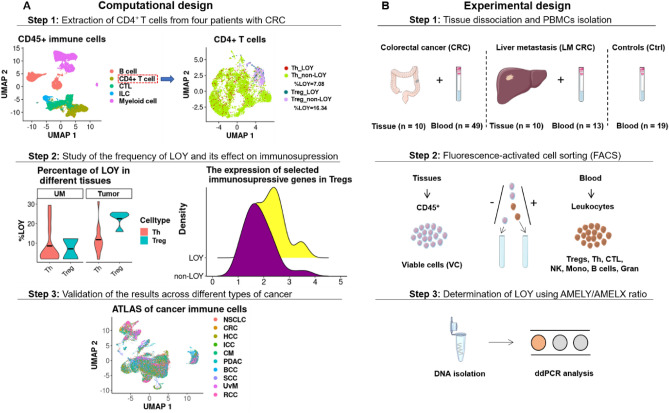
Figure 1.
Schematic workflow of computational (Panel A) and experimental (Panel B) design applied in our study. (A) Computational analysis of two scRNA-seq datasets; Zhang et al.32 and Nieto et al.33. In the first step, CD4 + T cells from the tumor microenvironment (TME), uninvolved margin (UM) and blood samples obtained from four CRC patients32 were re-clustered and categorized into Th and Tregs. Next, LOY-cells were defined based on the lack of expression of MSY-specific genes. Subsequently, the status of selected immunosuppressive genes was investigated in LOY and non-LOY Tregs located in the UM and tumor sites. The results obtained from the Zhang et al.32 dataset and comprising the scRNA-seq were validated in the Nieto et al.33 dataset, the latter covering an atlas of CD45 + immune cells from eight types of cancer, including CRC. (B) Experimental design for analysis of cells obtained from TME of human CRCs (n = 10) and LM_CRCs (n = 10) tissues and blood samples collected from CRC (n = 49), LM_CRC (n = 13) and Ctrl (n = 19) patients showed in three consecutive steps. Suspensions of single cells were sorted by FACS, followed by DNA isolation and ddPCR analysis using TaqMan-probes. Parts of the figure were drawn by using pictures from Servier Medical Art. Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License (https://creativecommons.org/licenses/by/3.0/). TME tumor microenvironment, UM non-tumorous uninvolved margin tissue, CRC colorectal cancer, LM_CRC liver metastasis of CRC, Ctrl control patients, Tregs regulatory T cells, Th helper T cells, CTL CD8 + cytotoxic T cells, ddPCR droplet digital PCR, scRNA-seq single-cell RNA-seq.