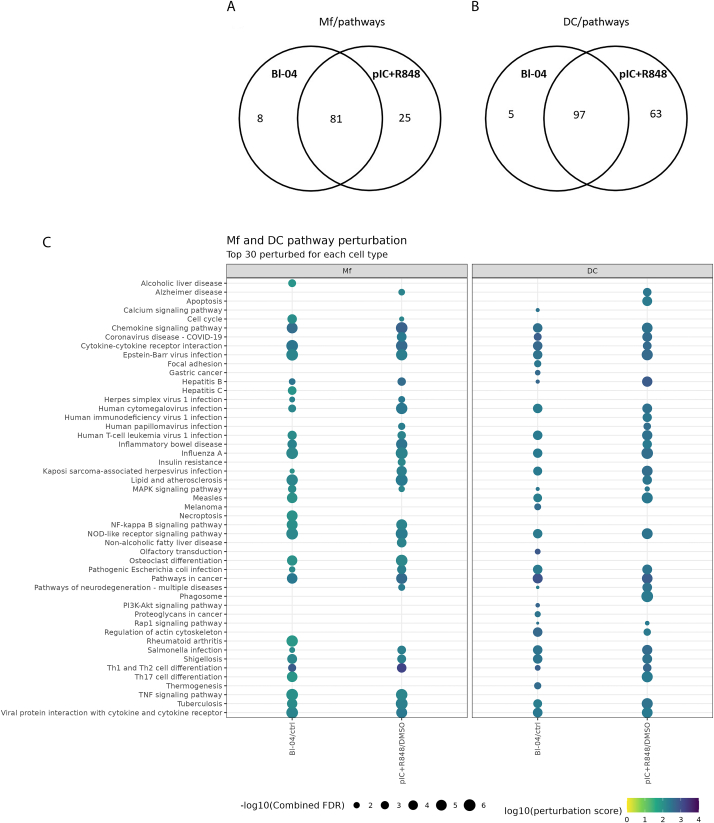
Fig. 3.
Pathway perturbation in immune cells. Differentially expressed gene results were used as input to ROntoTools to determine pathways that were affected by the treatments. The number of perturbed pathways for each treatment are shown for Mfs (A) and DCs (B). The top 30 most perturbed pathways are presented, for Mfs and DCs (C). The perturbation score is shown as the color (dark blue = higher values), and the size of the balloon denotes the –log10 of the combined false discovery rate (FDR). Only pathways that have FDR <0.05 (-log10 > 5) are shown. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)