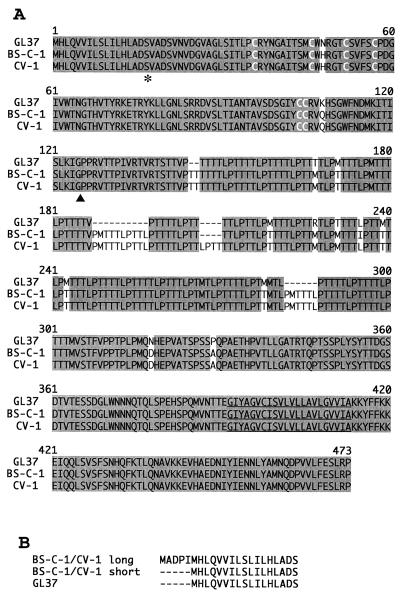
FIG. 3.
Alignment of havcr-1 from different AGMK cell lines. (A) Alignment of amino acid sequences predicted from the HAVcr-1 cDNAs of BS-C-1, CV-1, and GL37 cells was done with the Clustal W program. Gaps introduced in the sequences for the alignment are indicated by dashes; numbers of residues starting with the respective initiating methionine codons are indicated. Shading, identical amino acids; white letters, the six Cys residues of the Cys-rich region; asterisk, the end of the signal sequence; arrowhead, the beginning of the mucin-like region; underlining, the transmembrane region. (B) Alignment of the putative signal sequences predicted from the short and long forms of the BS-C-1 and CV-1 HAVcr-1 cDNAs compared to the GL37 havcr-1 signal sequence. The dashes indicate untranslated sequences. The amino acid sequences were inferred from the HAVcr-1 cDNA nucleotide sequences deposited in GenBank (see text).