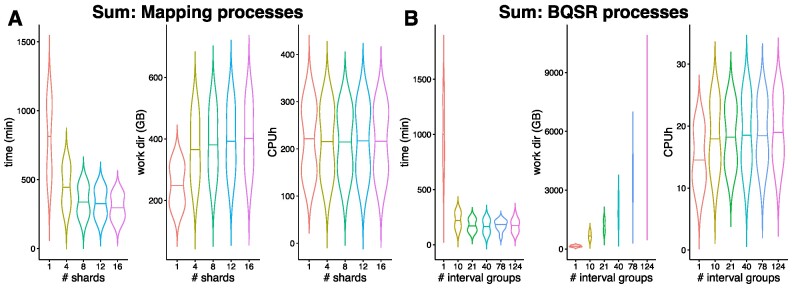
Figure 4.
Sharding the input FastQ files and parallelizing computation on interval groups reduces the overall runtime of the nf-core/sarek pipeline. (A) Effect of sharding the input files on the mapping processes, including fastP, BWA-MEM and Markduplicates. The input FastQ files were split into smaller pieces increasing the amounts of shards and the runtime, work directory size and CPU hours were evaluated for each split size. FastP was run with a different number of CPUs corresponding to the desired number of shards. (B) Effect of parallelizing computations across interval groups on BQSR processes, which include the BaseRecalibrator, GatherBQSRReports, ApplyBQSR and SAMtools merge process. When all intervals were processed together as one group the memory requests for ApplyBQSR had to be increased. The violin plots show computations on tumor-normal paired samples of five patients. The time was evaluated by summing up the highest realtime per task per sample as reported by the Nextflow trace report. The work directory size and CPU hours are the sums of all involved tasks.