Fig. 5. CRISPR/Cas9-RNP KO reveals that BATF regulates human activated Tregs by multiple signaling pathways.
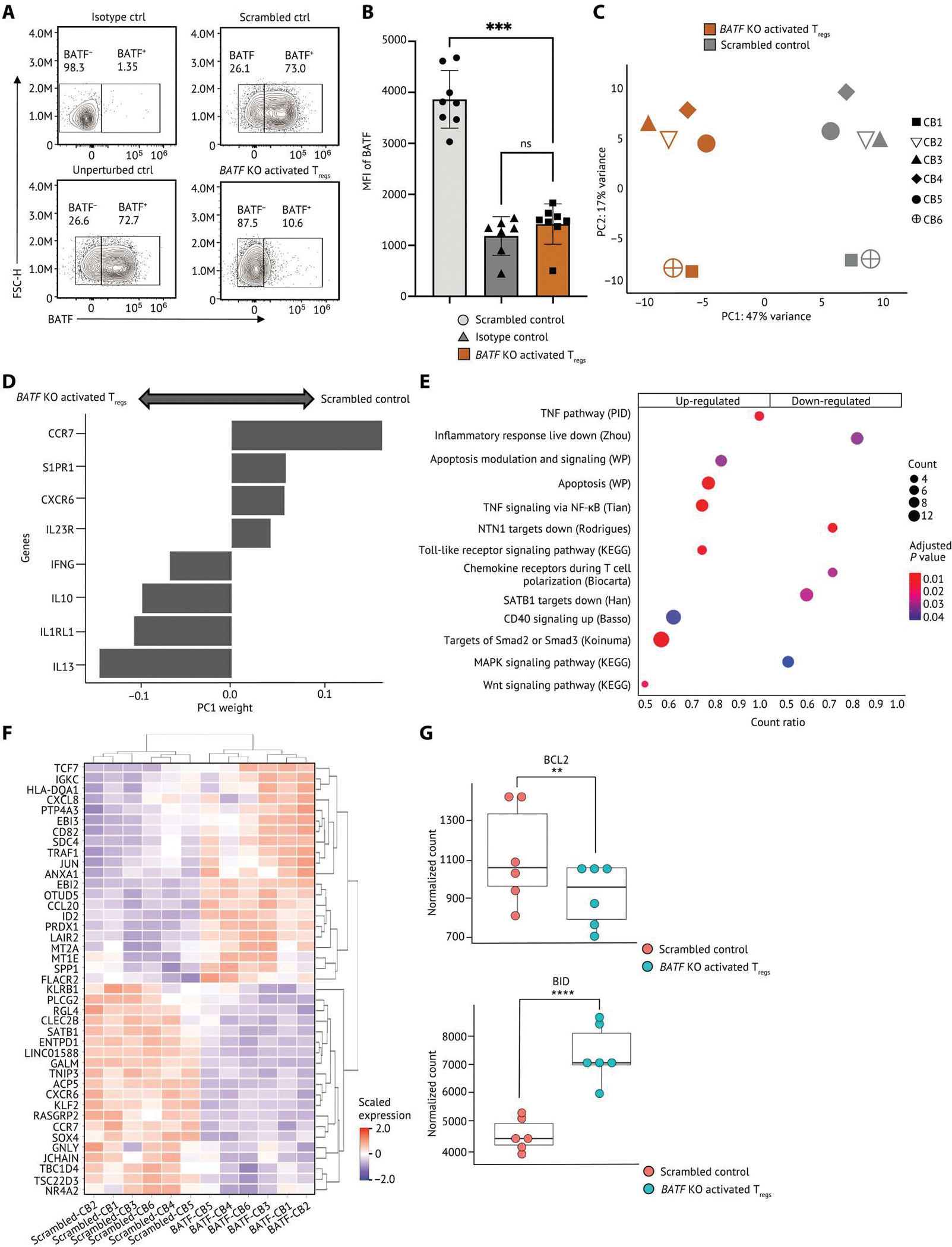
(A and B) Human primary Tregs isolated from cord blood were CRISPR-edited and then cultured with TCR stimulation for 3 days. (A) Representative flow staining of Tregs showing BATF protein level in nontargeting scrambled control and unperturbed control Tregs. (B) The BATF expression by median fluorescence intensity (MFI) in Tregs (n = 8) is shown. Each dot indicates an individual replicate. Bars indicate the median of expression, and error bars represent 1 SD. (C to G) RNA-seq was conducted on BATF KO activated Tregs (n = 6) and scrambled control from the same donor. (C) PCA of the transcriptome of the scrambled control and BATF KO activated Tregs. Human activated Tregs were stratified by PC1. (D) Weightings of the genes that were the strongest drivers of PC1 were visualized on a bar plot. (E) Selected gene sets that were enriched in human BATF KO activated Tregs were visualized on a dot plot. Genes are differentially expressed in both TNFR+ Tregs from scRNA-seq and BATF KO activated Tregs in bulk RNA-seq selected as input to the gene set enrichment analysis. Dots are colored by the false discovery rate (FDR)–adjusted P value, and dot sizes are scaled by the number of significantly up-regulated genes within each gene set (<0.1% FDR). (F) The relative expression of the top 20 up/down-regulated genes differentially expressed in both TNFR+ Tregs and BATF KO activated Tregs were visualized on a heatmap. The gene expression is scaled by row. (G) Dot plots showing the expression levels of BCL2 and BID in BATF KO activated Tregs and scrambled controls from eight individual replicates. Data in (B) were analyzed by a ratio paired t test, and the FDR P values in (G) were calculated by Wald test. P values: **P < 0.01; ***P < 0.001, ****P < 0.0001. ns, not significant.
