Fig. 2 |. Long non-coding RNAs as cis-acting transcription stabilizers.
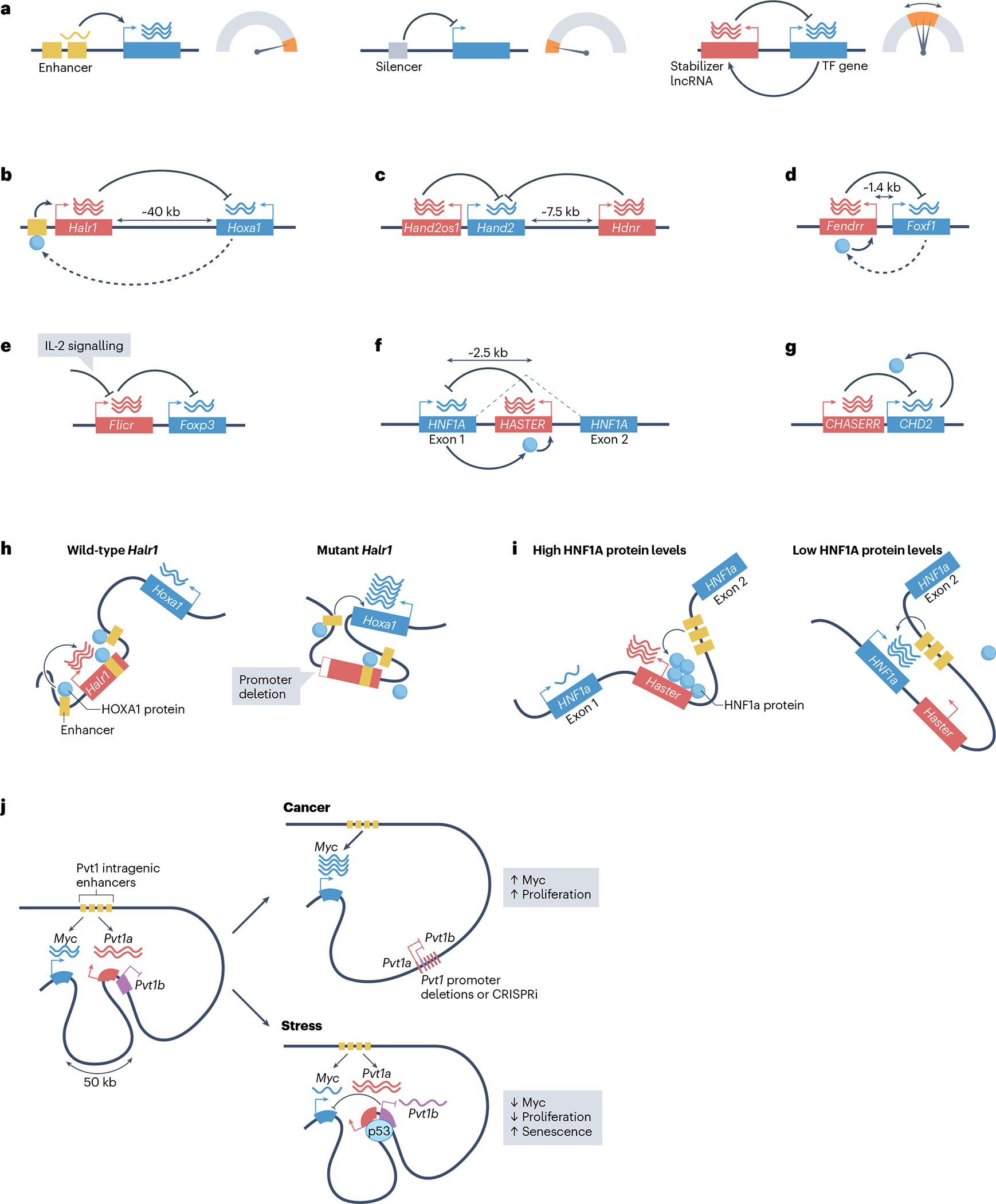
a, Whereas enhancers promote cell type-specific gene activation and silencers prevent the expression of their target genes, transcription-stabilizing long non-coding RNAs (lncRNAs) act in cis to tune the transcription level of dosage-sensitive transcription-factor (TF) genes. b–g, Examples of lncRNAs (in red) that act as transcription stabilizers of adjacent genes, all of which encode transcription regulators (in blue). The lncRNAs are Halr1 (b), Hand2os1 (c), FENDRR (d), Flicr (e), HASTER (f) and CHASERR (g) — loss of function of all of these lncRNAs caused increased expression of the adjacent gene. HNF1 homeobox A (HNF1A) and chromodomain helicase DNA-binding protein 2 (CHD2) (blue circles) enhance the inhibitory effects of the adjacent lncRNAs, and therefore provide negative feedback. Homeobox A (HOXA1) and forkhead box F1 (FOXF1) proteins are positive regulators of the lncRNAs Halr1 and FENDRR, respectively, suggesting they could also form a negative feedback loop. Some transcription-stabilizing lncRNAs modulate their target genes in a signal-responsive manner; for example, interleukin-2 (IL-2) acts on Flicr (e) to reduce high Foxp3 expression levels in regulatory T cells. Two lncRNAs, Hand2os1 and Hdnr (c), restrict Hand2 expression. h,i, The promoters of transcription-stabilizing lncRNAs modulate interactions between their target TFs genes and local enhancers. h, Left: in pluripotent cells, Halr1 binds and sequesters proximal enhancers of Hoxa1, which dampens retinoic acid-induced expression of Hoxa1. Right: deletion of the Halr1 promoter increases enhancer–Hoxa1 interactions. HOXA1 (blue circles) binds to local enhancers and activates Halr1, which restrains Hoxa1 expression. Left and right in h depict retinoic acid-induced cells. i, The Haster promoter limits interactions between the Hnf1a promoter and intragenic enhancers. This effect is accentuated at high concentrations of HNF1A protein, thereby providing negative feedback on Hnf1a transcription. j, The active Pvt1 lncRNA promoter acts as a boundary element that associates with enhancers located within the Pvt1 gene body and limits access of the Myc promoter to these enhancers. Experimental inhibition of the transcription activity of the Pvt1 promoter through targeted promoter deletions or CRISPR inactivation (CRISPRi) leads to increased Myc promoter–enhancer engagement, high Myc transcription and increased cellular proliferation. The Pvt1 locus also harbours a p53-dependent isoform, Pvt1b, which downregulates Myc transcription during stress, decreases cell proliferation and increases cell senescence without apparent changes in Myc–enhancer contacts.
