Figure 3. Metabolic features of the NADH reductive stress in HeLa cells.
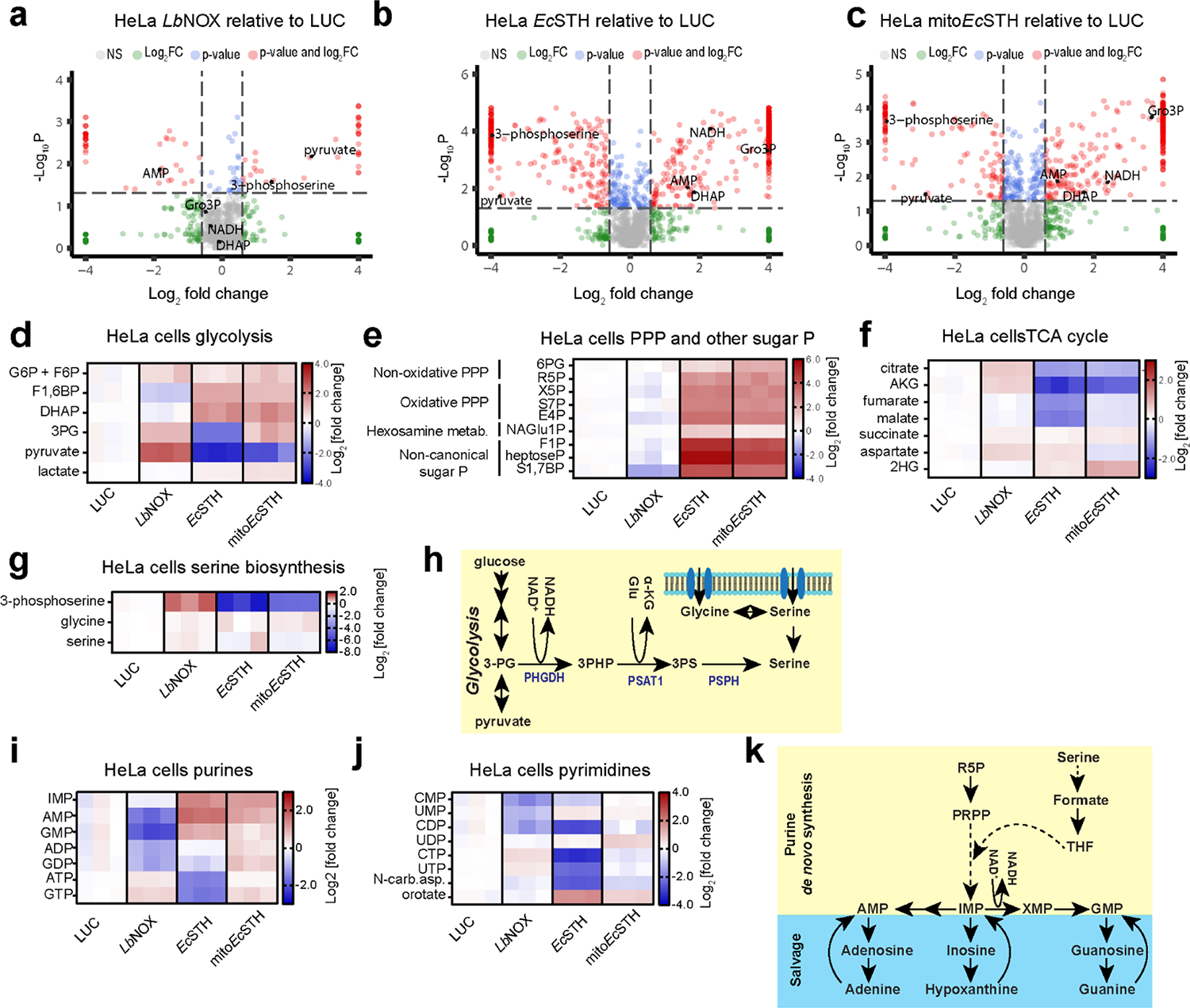
(a-c) Untargeted metabolomics of HeLa cells expressing LbNOX, EcSTH and mitoEcSTH. Heatmaps of the most impacted glycolysis (d), pentose phosphate pathway, non-canonical sugar phosphates (e), the TCA cycle (f), serine biosynthesis (g) intermediates in HeLa cells expressing EcSTH and mitoEcSTH. G6P: glucose-6-phosphate; F6P: fructose-6-phosphate; F1,6BP: fructose-1,6-biphosphate; DHAP: dihydroxyacetone phosphate; 3PG: 3-phosphoglycerate; 6PG: 6-phosphogluconate; R5P: ribose-5-phosphate; X5P: xylulose-5-phosphate; S7P: sedoheptulose-7-phosphate, E4P: erythrose-4-phosphate; NAGlu1P: N-acetylglucosamine-1-phosphate; F1P: fructose-1-phosphate; heptoseP: a putative heptose with one phosphate; S1,7BP: sedoheptulose-1,7-bisphosphate; AKG: α-ketoglutarate; 2HG: 2-hydroxyglutarate. (h) Schematic of serine cellular biosynthesis. Heatmaps of the most impacted purines (i) and pyrimidines (j) in EcSTH and mitoEcSTH expressing HeLa cells. N-car.asp.: N-carbamoyl-aspartate. (k) Schematic of purine cellular salvage and biosynthesis. Luciferase and LbNOX expressing HeLa cells were used as controls in (a-g, i, j). In heatmaps (d-g, i, j) each column represents biologically independent sample. The statistical significance indicated for (a-c) represents p value cutoff = 0.05, fold change cutoff = 1.5 and Welch t test (FDR corrected).
