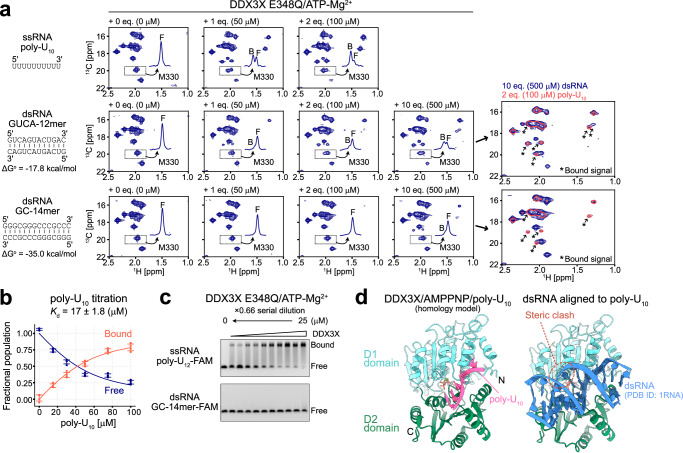
Fig. 3. RNA titration experiments.
a 13C-1H HMQC spectra of [Frac-2H; Ileδ1-13C1H3; Metε-13C1H3]-labeled DDX3X E348Q in the presence of varying concentrations of poly-U10 ssRNA (top), GUCA-12mer dsRNA (middle), or GC-14mer dsRNA (bottom) are shown. The overlay of the spectra of DDX3X measured with 2 equimolar poly-U10 (coral, single contour) or 10 equimolar dsRNA (navy, multiple contours) are also shown for GUCA-12mer and GC-14mer dsRNA datasets. The newly appeared bound state signals are highlighted by arrows. Duplex stability (ΔGo) at 35 °C was estimated by using nearest-neighbor parameters in 1 M NaCl49,50. b Plots of the fractional populations of the free (navy) and bound (orange-red) states as a function of poly-U10 concentration. Experimental data points are shown as circles, and the fitted curve is shown as a line. Error bars represent the standard deviation of the data obtained from four different methyl correlations. The center of the error bar represents the average value. c EMSA binding experiments for DDX3X using poly-U12-FAM ssRNA (top) or GC-14mer-FAM dsRNA (bottom). DDX3X E348Q variant protein was titrated from 0 to 25 μM. The titration was carried out in the presence of 5 mM ATP/MgCl2. The binding experiments using poly-U12-FAM ssRNA were repeated three times with similar results, while the binding experiments using GC-14mer-FAM dsRNA were repeated twice with similar results. d Model structure of DDX3X bound to AMPPNP and poly-U10 (left) or 14mer dsRNA (PDB ID: 1RNA)82 (right). All NMR measurements were performed at 35 °C and 600 MHz, and the protein concentration was 50 μM. The 1D projections of the dotted region containing the free (F) and bound (B) signals for M330 are shown in each spectrum. Source data are provided as a Source Data file.