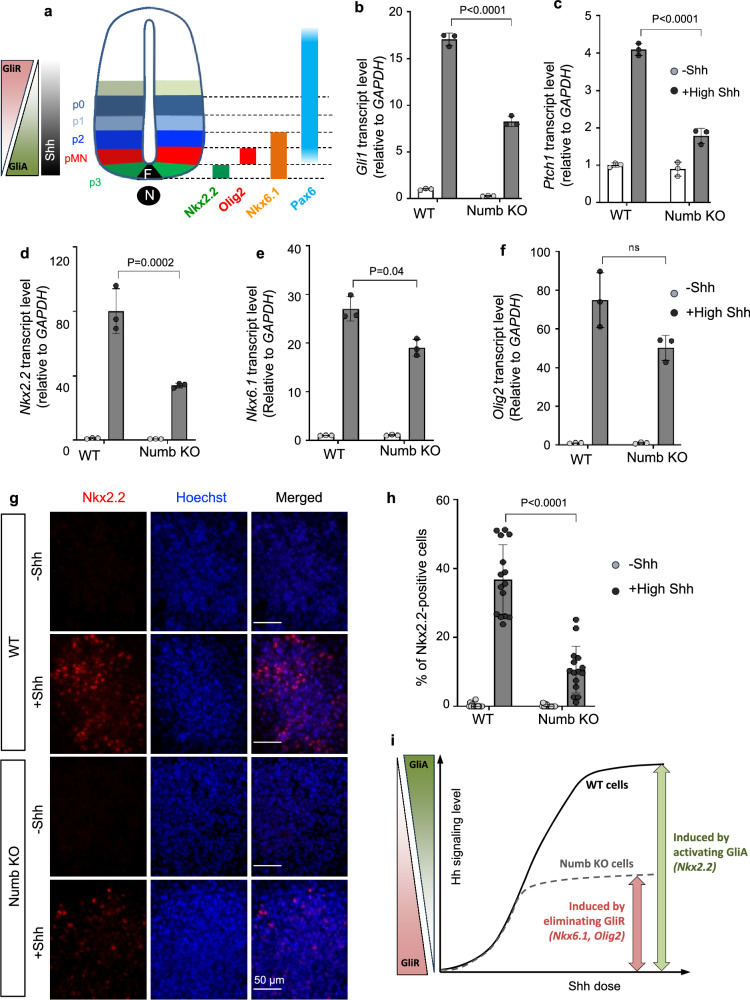
Fig. 7. Numb is required for high-level Hh responses in mouse spinal cord neural progenitor cells (NPCs).
a Diagram illustrating embryonic spinal cord progenitors and their associated markers (adapted from Ribes et al. 49). The bars on the right side depict the specific transcription factors in their respective domains. N notochord, FP floor plate, pMN motor neuron progenitors; p0, p1, p2, and p3, ventral interneuron progenitors. b, c Hh signaling activity was assessed by the transcript levels of Hh-target genes, Gli1 and Ptch1, in WT and Numb KO mouse spinal cord NPCs. NPCs underwent differentiation in two conditions: either retinoic acid (RA) without Shh, or RA with 100 nM Shh (+High Shh) for 3 days. d–f Transcription levels of specific transcription factors induced by high Shh in WT and Numb KO NPCs. Cells were differentiated in RA-containing medium without or with 100 nM Shh for 3 days. Three independent differentiation experiments were performed with similar results. n = 3 biological replicates. Statistics in (b–f): Two-way ANOVA with multiple comparisons (Tukey test). Data are shown as mean ± SD. g Representative images of NPC differentiation into Nkx2.2-positive NPCs after 3 days of induction. h The percentage of cells positive for Nkx2.2 in WT or Numb KO NPCs. n = 15 images from three independent experiments were quantified for each condition. Each image represents one NPC colony consisting of approximately 200–300 cells. Data are shown as mean ± SD. Statistics: Two-way ANOVA with multiple comparisons (Tukey test). i Schematics of Hh signaling and NPC differentiation. NPC cell fate in the spinal cord is ultimately determined by the opposing gradient of Gli activator (GliA) and Gli repressor (GliR). In Numb-null ES cells, even with high Hh stimulation, GliA activation is impeded, but cells are still able to curb Gli3R production. Consequently, Numb-null ES cells fail to differentiate into cell fates that depend on GliA (Nkx2.2-positive NPC) but are able to differentiate into cell fates that are reliant on Gli3R elimination (Olig2- or Nkx6.1-positive NPCs). Source data are provided as a Source Data file.