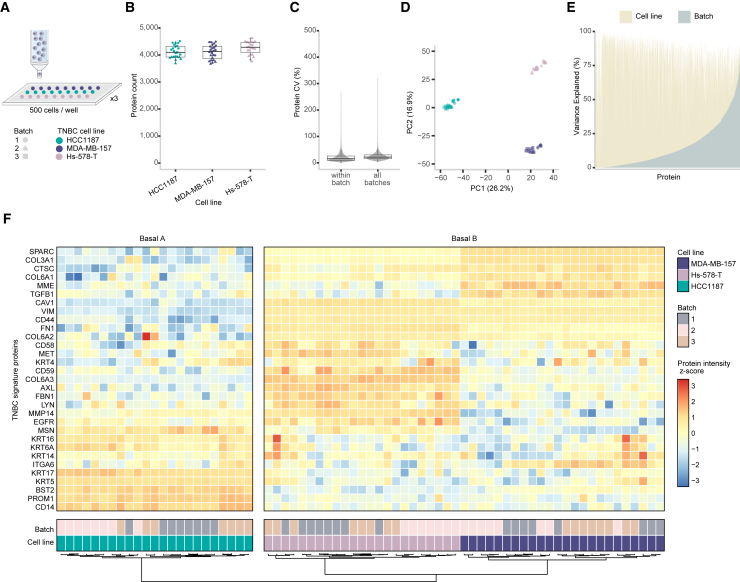
Figure 2.
Evaluation of DROPPS for profiling TNBC cell line differences across batches
(A) Workflow for testing inter-batch variation by DROPPS. Sorted cells (500 cells) from three TNBC cell lines were processed separately on different days. Each cell line had seven or eight sample replicates processed each day.
(B) The protein counts of the individual runs.
(C) Protein CVs for runs within a batch or for combining batches.
(D) First two principal components of PCA.
(E) The percentage of variance explained by cell line and batch from two-way ANOVA (i.e., omega-squared).
(F) Z scored log2(intensity) of TNBC signature proteins21 using data acquired by DROPPS.
Boxplots show the median, interquartile ranges, and 95% confidence interval estimate. TNBC, triple negative breast cancer.