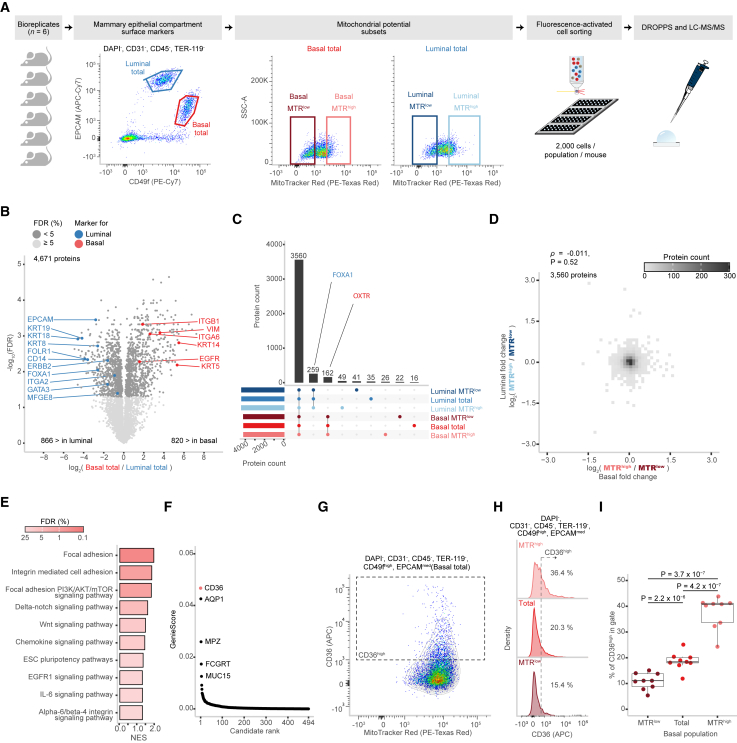
Figure 3.
Proteomic profiling of primary mammary epithelial cells sorted by mitochondrial potential
(A) Schematic of experimental design and FACS gating scheme used to sort subsets of mammary epithelial cells of basal and luminal compartments based on their mitochondrial potential as marked by MTR dye (n = 6 each) that were subsequently processed using DROPPS.
(B) Differences in log2(intensities) of proteins from the total basal and total luminal populations. Points are colored by significance, and known compartment markers are labeled.
(C) Overlap in proteins detected between select subsets of epithelial cells.
(D) 2D bin plot depicting the distribution of log2 fold changes between MTRhigh and MTRlow for the basal and luminal compartments. Spearman correlation and associated p value are shown.
(E) Select gene set enrichment analysis (GSEA)-enriched pathways in the MTRhigh population compared to the total population for the basal and luminal compartments.
(F) Rank plot depicting the distribution of GenieScores for proteins with surface prediction consensus >0 calculated using SurfaceGenie26 on mean log2(intensities).
(G) Representative scatterplot depicting the relationship between CD36 and MTR fluorescent intensity.
(H) Representative histograms depicting the distribution of CD36 flow cytometry signal in MTRlow, total, and MTRhigh basal populations.
(I) The percentage of CD36high cells in different MTR fractions of basal cells shown with paired t test p values (n = 8).
Boxplots show the median, interquartile ranges, and 95% confidence interval estimate. MTR, MitoTracker Red.