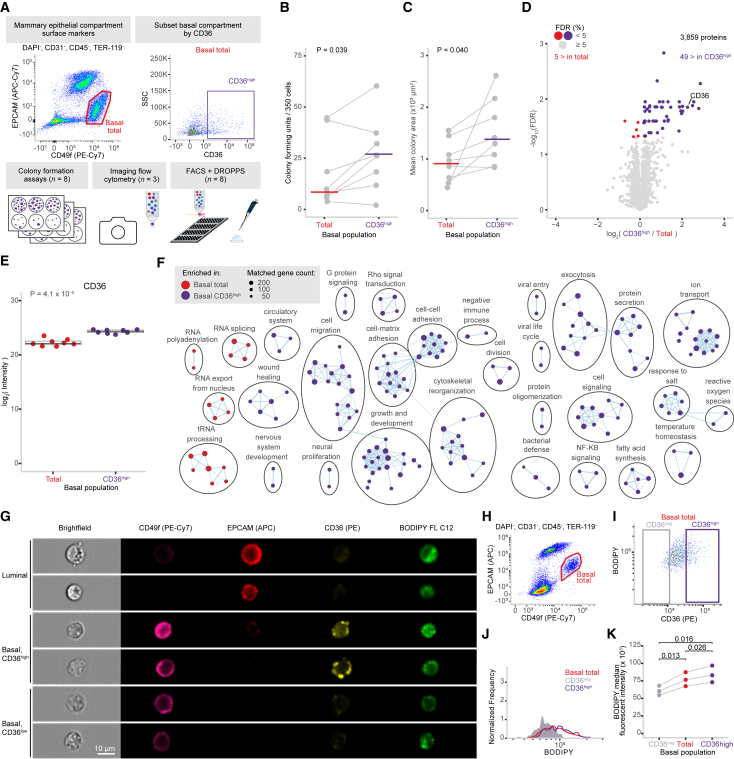
Figure 4.
Investigating the phenotype of CD36high basal mammary epithelial cells
(A) Schematic of experimental design and FACS gating scheme used to sort subsets of mammary epithelial cells of basal and luminal compartments based on CD36 expression that were subsequently used for CFC, processed using DROPPS, or analyzed by imaging flow cytometry.
(B and C) Line plots depicting the (B) number and (C) size of the colonies formed by basal cells sorted by CD36 expression shown with median value and paired t test p values.
(D) Differential protein expression results from total basal and CD36high basal populations. Points are colored by significance and directionality.
(E) The log2(intensity) of CD36 from individual runs shown with the paired t test p value (n = 8).
(F) Cytoscape enrichment map for significantly enriched GO Biological Processes between total basal and CD36high basal populations.
(G) Representative images from imaging cytometry highlighting the surface markers used to distinguish epithelial compartments along with CD36 and the fluorescent fatty acid analog BODIPY.
(H) Scatterplot depicting the gating scheme used to identify basal epithelial cells.
(I) Scatterplot depicting the relationship between BODIPY and CD36 intensities along with CD36neg and CD36high gates.
(J) Representative histogram depicting the BODIPY intensity distributions among CD36neg, total, and CD36high basal populations.
(K) Dot plot depicting the median BODIPY fluorescent intensity in CD36neg, total, and CD36high basal populations along with paired t test p values.
CFC, colony-forming capacity.