Fig. 2.
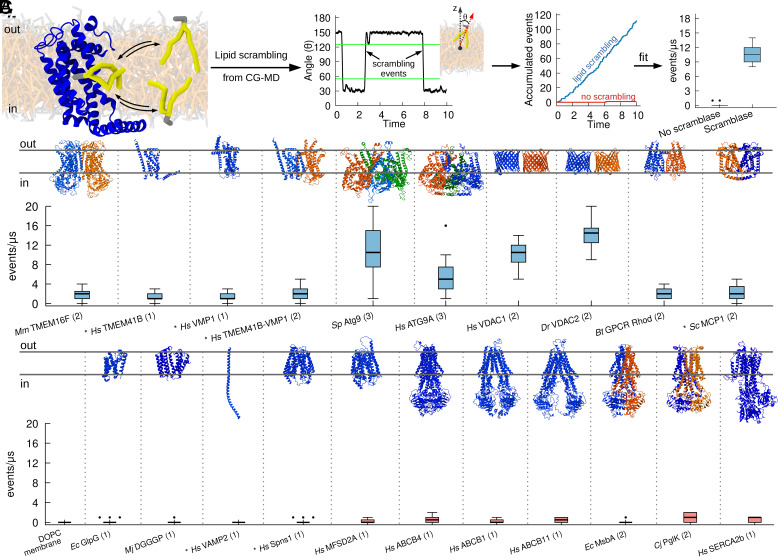
CG simulations recapitulate known activity of lipid scramblases. (A) Protocol used to quantify lipid scrambling in CG-MD simulations. For denoising purposes, the time traces for each final angle were smoothened by averaging over a 50-ns window and data points were collected each 25 ns. A scrambling event was counted when a lipid in the upper leaflet moved to an angle greater than 125°, and when a lipid in the bottom leaflet moved to an angle lower than 55°. These thresholds are highlighted by green lines. (B) CG-MD simulations reproduce lipid scrambling activity by known lipid scramblases of different structures and oligomerization states. (C) CG-MD simulations correctly reproduce lack of lipid scrambling activity by proteins that do not have scrambling activity in vitro. AlphaFold structures are denoted by the * symbol, oligomerization state is in parenthesis.