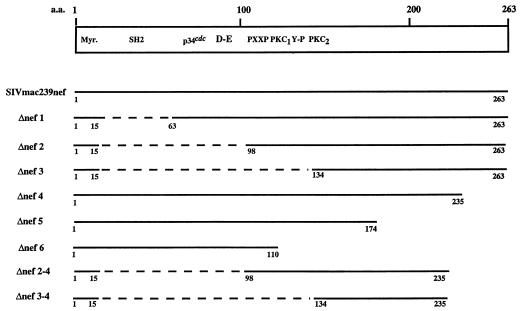
FIG. 1.
Schematic representation of deletion mutations in nef of SIVmac239. Sequence motifs are described by Shugars et al. (53) and Samuel et al. (49). Myr, myristoylation site; SH2, putative consensus SH2 binding sequence; p34cdc, consensus cyclin-dependent kinase substrate sequence; D-E, acidic stretch; PXXP, putative SH3 binding motif; Y-P, tyrosine kinase recognition sequence; PKC1 and PKC2, PKC recognition sequences. Clones for expression in mammalian cells contained an AU1 epitope tag at the carboxyl termini. Dashed lines represent deleted regions; numbers indicate the positions of the amino acids.