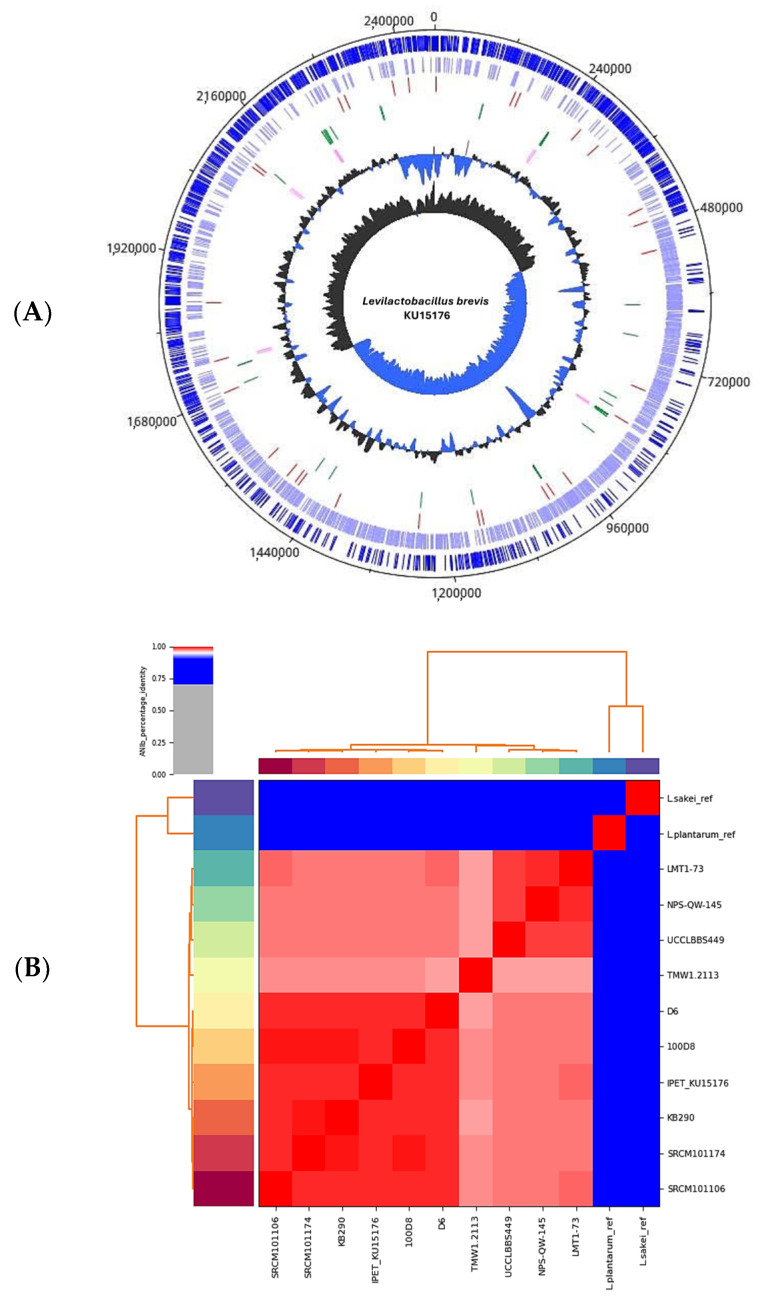
Figure 1.
Genome analysis of L. brevis KU15176. (A) The genome map and features of L. brevis KU15176. From the outside going in, it includes CDS in the forward strand, CDS in the reverse strand, miscellaneous RNA, transfer RNA, ribosomal RNA, transfer-messenger RNA, GC plot, and GC skew. The genome map was generated using DNAPlotter implemented in Artemis (v.18.2.0) while the features were provided by QUAST (v.5.2.0) and Prokka (v1.14.6). (B) Percentage identity heatmap of L. brevis KU15176 against reference L. brevis strains (LMT1-73, NPS-QW-145, UCCLBBS449, TMW1.2113, D6, 100D8, KB290, SRCM101174, SRCM101106) and L. plantarum SK151 (L.plantarum-ref) and L. sakei CBA3614 (L.sakei-ref) strains as outliers verified the identity of L. brevis KU15176 that is >95% similar to that of the reference L. brevis strains. The heatmap was generated by implementing the ANIb method in pyani (v0.2.12).