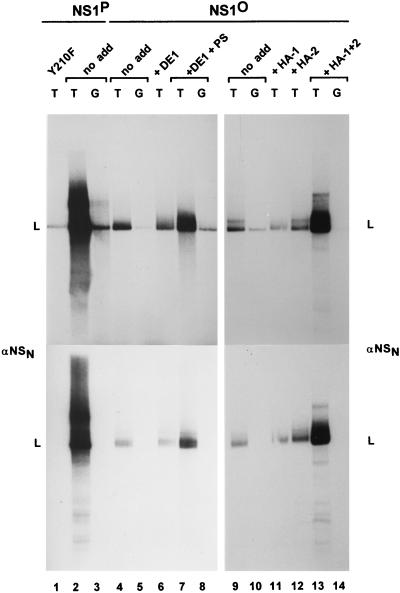
FIG. 5.
Activation of NS1O for RCR by fractionated HeLa cell extracts. NS1-dependent RCR of plasmids containing the left-end active (T) or inactive (G) origin was determined in a kinase-free in vitro system based on P1-Thr and T4 DNA polymerase. Activation of NS1O (lanes 4 to 14) was achieved by using the indicated protein components (see Fig. 4) in the presence or absence of protein kinase C cofactors. Only some of the protein-cofactor combinations that have been tested are illustrated, as most representative. Lanes 4 to 8 show the dependency of NS1O activation on PKC cofactors. PS, Ca2+ plus PS. Lanes 9 to 14 show the segregation of protein components allowing the reactivation of dephosphorylated NS1. The illustrated reactions were carried out in presence of the PKC activators Ca2+ and PS (HA-1) or TPA (HA-2 and HA-1 plus HA-2). It should be mentioned that TPA-stimulated HA-1 or Ca2++PS-stimulated HA-2 also failed to activate NS1O (data not shown), while the combined protein components (HA-1 plus HA-2) were able to rescue NS1O in the presence of Ca2++PS instead of TPA. The NS1 mutant Y210F served as a negative control (lane 1). Native NS1P was used as a phosphorylated, replication-competent standard (lanes 2 and 3). L, migration of the linearized plasmid. The lower panel presents reaction products after immunoprecipitation with αNSN.