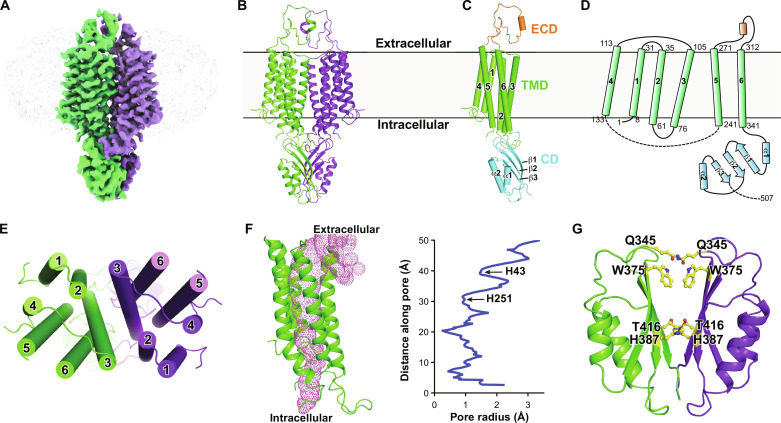
Fig. 2. Overall architecture of hZnT1.
(A) EM density for the hZnT1-D47N/D255N double mutant (hZnT1-DM) at 3.4-Å resolution. Densities corresponding to the two hZnT1 protomers are colored green and purple, respectively. The surrounding micelle is shown in light gray. (B) Cartoon representation of the dimeric hZnT1-DM structure. (C) A side view of the hZnT1-DM protomer structure, which is organized into the TMD, CD, and ECD. (D) Topological diagram of hZnT1. The starting and ending residue numbers of each transmembrane helix are labeled aside. (E) A top view of the TMD of hZnT1-DM structure. (F) Ion accessibility analysis of the hZnT1 structure, as calculated using HOLE2 (49). The pore cavity is shown as magenta mesh. Calculated pore radius along the transmembrane region is shown. (G) Overview of the CD of hZnT1-DM. Residues in the dimer interface forming either hydrophobic interaction or hydrogen bonding are shown in sticks.