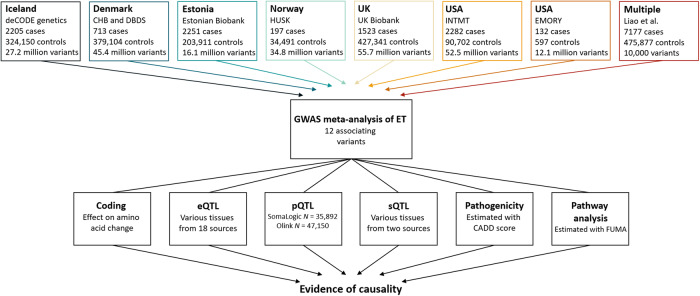
Fig. 1. Study design.
The first row lists the datasets used in the GWAS meta-analysis, number of ET cases, controls and variants analyzed. We included variants with MAF > 0.01% in all datasets except for the Estonian dataset and the previous GWAS12, where variants with MAF > 1% were included. The summary data from a previous GWAS, only includes the top 10,000 variants. The last row lists the multiomics approaches used to search for potential causal genes. Expression quantitative trait loci (eQTL) data sources are listed in Supplementary Data 15. Plasma protein levels (pQTL) were measured in Icelandic samples using Somalogic platform and in UK samples using the Olink platform. Splicing quantitative loci (sQTL) data were estimated using Icelandic RNA sequencing data, in addition to data imported from GTEx.