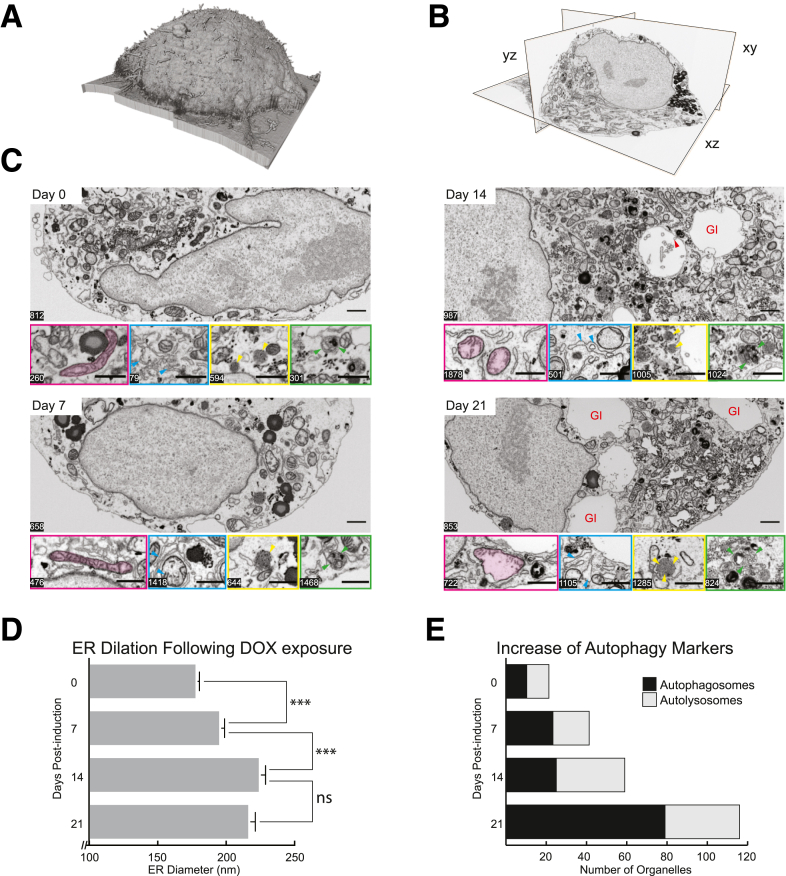
Figure 9.
Nanotomographic analysis of HepG2 cells at different time points postinduction of ATZ expression. (A) Volume rendering and (B) orthogonal view, showing a 21-day postinduction cell in xy, yz, and xz planes. Note the ability to reconstruct the data set in any axis, afforded by the isometric voxels. (C) Representative single-plane images from each of the 4 time points assayed. Inset panels highlight changes in organelle ultrastructure at different z-slice positions, such as increase in ER diameter and the density of autophagic organelle occurence (magenta--mitochondria, cyan--endoplasmic reticulum, yellow--autophagosomes, and green--autolysosomes). Cyan arrowheads illustrate the increased dilation of endoplasmic reticulum over the time course. Yellow arrows indicate the presence of lipid droplets Green arrowheads indicate the presence of autophagosomes. Red arrowhead in day 14 illustrates the presence of omegasomes in the emergent globular inclusions (as denoted by the red GI label). Scale bar: 1 μm. (D) Quantification of the increase in the mean diameter of the endoplasmic reticulum per cell at each of the 4 time points. ∗∗∗ denotes P < .001 (E) Increased density of autophagosomic organelles, as measured by the mean number of autophagosomes and autolysosomes counted per cell at each of the 4 time points. GI, globular inclusions.