Abstract
Objective
Metamorphosis is a transition from growth to reproduction, through which an animal adopts adult behavior and metabolism. Yet the neural mechanisms underlying the switch are unclear. Here we report that neuronal E93, a transcription factor essential for metamorphosis, regulates the adult metabolism, physiology, and behavior in Drosophila melanogaster.
Methods
To find new neuronal regulators of metabolism, we performed a targeted RNAi-based screen of 70 Drosophila orthologs of the mammalian genes enriched in ventromedial hypothalamus (VMH). Once E93 was identified from the screen, we characterized changes in physiology and behavior when neuronal expression of E93 is knocked down. To identify the neurons where E93 acts, we performed an additional screen targeting subsets of neurons or endocrine cells.
Results
E93 is required to control appetite, metabolism, exercise endurance, and circadian rhythms. The diverse phenotypes caused by pan-neuronal knockdown of E93, including obesity, exercise intolerance and circadian disruption, can all be phenocopied by knockdown of E93 specifically in either GABA or MIP neurons, suggesting these neurons are key sites of E93 action. Knockdown of the Ecdysone Receptor specifically in MIP neurons partially phenocopies the MIP neuron-specific knockdown of E93, suggesting the steroid signal coordinates adult metabolism via E93 and a neuropeptidergic signal. Finally, E93 expression in GABA and MIP neurons also serves as a key switch for the adaptation to adult behavior, as animals with reduced expression of E93 in the two subsets of neurons exhibit reduced reproductive activity.
Conclusions
Our study reveals that E93 is a new monogenic factor essential for metabolic, physiological, and behavioral adaptation from larval behavior to adult behavior.
Keywords: Monogenic factor of obesity, Systemic metabolic failure, Exercise endurance, Circadian rhythm, Neuronal regulation of feeding, Brain rewiring, Adult behavior
Highlights
-
•
A screen of 70 Drosophila homologs enriched in mammalian VMH identified E93, a homolog of mammalian LCOR and LCORL.
-
•
Neuron-specific reduction of E93 expression results inhyperphagia, obesity, and impaired exercise ability and circadian rhythm.
-
•
These defects were because of E93 action mainly in two types of neurons: GABA-ergic and MIP-producing neurons.
-
•
The systemic metabolic defects could be caused by the defects in brain rewiring during adolescence transition.
1. Introduction
The nervous system integrates environmental and internal cues and generates appropriate physiological and behavioral responses for animals to survive unpredictable and dynamic environments. This physiological and behavioral adaptation often requires a switch between different metabolic states, as best exemplified in insects that go through metamorphosis. Entirely focused on growth, larvae feed constantly to fulfill their metabolic demand, such that their metabolism relies on aerobic glycolysis, similar to cancer metabolism [[1], [2], [3]]. After metamorphosis, however, the focus of an adult switches to reproduction, where the adults execute diverse behavioral paradigms to seek mates, succeed in mating, and produce progeny. With these multiple tasks to attend, the importance of feeding diminishes in adults. Indeed, many species of moths, such as Actias luna, the American moon moth, and most of the Saturniidae family to which A. luna belongs, do not have a feeding organ in adults, and survive and reproduce solely on the nutrients they preserved as larvae [4].
In Drosophila, the switch between growth and metamorphosis is mediated by two essential endocrine signals, Juvenile Hormone (JH) and Ecdysone (20E, a steroid hormone derivative of cholesterol) [5]. JH promotes growth, antagonizing 20E action in metamorphosis, and ensures that larvae grow big enough before they become adults. The balance between these two hormones, which is tightly controlled by the animal's metabolic state and nutritional environment, determines the timing of development and metamorphosis [[6], [7], [8]]. Metamorphosis itself is a prolonged starvation for over 3 days, and certain metabolically-challenged mutants cannot survive it [1]. In adults, JH and 20E regulate reproduction which also requires an on-off switch upon the females' mating status, suggesting their continuous roles in balancing metabolic states after metamorphosis.
From an RNA interference-based screen designed to discover novel yet conserved metabolic regulators, we identified E93 (Eip93F: Ecdysone-induced protein 93F), an ortholog of mammalian ligand dependent nuclear receptor corepressor (LCoR), and ligand dependent nuclear receptor corepressor-like (LCoRL). In the screen, 70 Drosophila orthologs to the mammalian genes enriched in ventromedial hypothalamus (VMH) were selected, individually knocked down in a neuron-specific manner then screened for starvation resistance. We focused on the genes enriched in VMH based on its essential roles in metabolic homeostasis [9].
E93, a master regulator of metamorphosis, belongs to the Pipsqueak transcription factor family [10,11] and contains multiple functional domains such as the DNA binding Helix-Turn-Helix (HTH) domain, the CtBP-interacting domain (CtBP-im) and two potential nuclear hormone interaction domains (NR-box) [12,13]. Conserved from Caenorhabditis elegans to mammals [14], it regulates neurite pruning in C. elegans [13] and metamorphosis in insects, including Drosophila [12]. A mammalian ortholog, LCORL, is associated with body size variations and lipid metabolism in mammals [15,16]. In Drosophila, E93 plays an essential role during metamorphosis to suppress expression of larval genes and promote expression of adult genes [17]. The enhancer sites targeted by E93 continuously change throughout metamorphosis, suggesting that dynamic interactions between E93 and stage-specific factors allow E93 to serve as a switch for the transition of development stages [18]. E93 is also required for termination of neurogenesis in mushroom body neuroblasts, presumably to help the neurons adopt their adult fate [19]. E93 expression is maintained at low levels until larva L3 puffstage, the beginning of metamorphosis, when expression is induced by ecdysone (20E) [20]. E93 expression persists in adults mostly in neurons, including subsets of antennal sense organ and olfactory neurons [21], suggesting its roles beyond metamorphosis. Despite its essential roles in this developmental program tightly linked to metabolic transition, whether neuronal E93 plays any role in adaption to adult metabolism, physiology, and behavior has not been studied. Furthermore, the important sites of E93 action for them are completely unknown.
Here we report that reduced expression of E93 in a neuron-specific manner results in hyperphagia and various metabolic and physiological abnormalities. Knockdown of E93 increases attraction to food, food intake, body weight, and energy stores with impaired exercise endurance and disrupted circadian rhythm. E93 knockdown in a specific developmental time window caused obesity, suggesting that E93 action during metamorphosis, the transition from larval phase to adult, is critical for regulation of adult metabolism. The systemic failure of metabolic homeostasis is largely due to E93's action in two populations of neurons: GABA-ergic and myoinhibitory peptide (MIP)-producing neurons (from here, MIP neurons), both of which have been implicated in controlling food intake, metabolism, circadian rhythm, and sleep [[22], [23], [24], [25], [26], [27], [28]]. MIP neuron-specific knockdown of the ecdysone receptor (EcR), the receptor of ecdysone (20E) which acts upstream of E93 in metamorphosis, partially phenocopies the obesity phenotype of MIP neuron-specific knockdown of E93. Additionally, restoring E93 only in MIP neurons partially reverses the metabolic abnormalities observed in neuron-specific knockdown of E93. Finally, the animals with reduced neuronal E93 expression fail to exhibit proper adult behavior, prioritizing feeding or resting over mating. Together, our study reveals neuronal E93 as a novel monogenic regulator in controlling adult metabolism, physiology, and behavior.
2. Materials and methods
2.1. Drosophila strains and growth conditions
The following lines were obtained from the Bloomington Drosophila Stock Center: nSyb-GAL4 (#51635), vGlut-GAL4 (#26160), ChAT-GAL4 (#6793), vGAT-GAL4 (#58409), Trh-GAL4 (#38389), Tdc2-GAL4 (#9313), OK107-GAL4 (#854), Akh-GAL4 (#25683), Mip-GAL4 (#51983), Ilp2-GAL4 (#37516), Eth-GAL4 (#51982), Fmrfa-GAL4 (#56837), Lk-GAL4 (#51993), NPF-GAL4 (#25681), phm-GAL4 (#80577), ple-GAL4 (#8848), SifAR-GAL4 (#76670), Tk-GAL4 (#51973), Eip93F RNAi (#57868), EcR RNAi (#58286), vGAT RNAi (#41958), and tubP-GAL80ts (#7019). E93-RNAi, nSyb-GAL4, mCherry-RNAi lines were outcrossed for five generations to a Berlin background (originally from the Heisenberg lab). Flies were raised on standard cornmeal/molasses food and incubated at 25 °C under a 12 h light/12 h dark cycle.
2.2. Quantification of triglycerides (TAG)
Triglycerides and protein were quantified as described by Tennessen et al. [29] with minor modifications. 5 flies were decapitated, collected into 1.5 ml tubes, and kept on dry ice before adding 50 μl of 0.5% PBS-T. They were then homogenized, heated for 5 min at 70 °C, and centrifuged at 14,000 rpm for 3 min at 4 °C. Resulting supernatant was transferred to a new 1.5 ml tube and placed on ice. 200 μl of Infinity Triglyceride reagent (ThermoFisher Scientific, Waltham, MA; catalog no. TR22421) were added to a 96 well plate. 4 μl of supernatant from each sample was transferred to each well in duplicates and the plate was incubated at 37 °C for 30–60 min. Absorbance levels were read at 540 nm.
2.3. Quantification of protein
Protein level was quantified using the Pierce BCA Protein Assay kit (ThermoFisher Scientific, Waltham, MA; product no. 23228) and used to normalize triacylglycerides (TAG) and glycogen levels. 4 μl of each sample (collected by the method described above) was added to each well of a 96 well plate. Volume of BCA reagent A was determined using the formula: (2 × number of samples + 20) × 100. BCA reagent A and B were mixed (A:B = 50:1) and 100 μl of this mixture was added to each well. The plate was incubated at 37 °C for 30 min and absorbance values were read at 562 nm of wavelength.
2.4. Quantification of glycogen
Glycogen quantification was performed as described by Tennessen et al. [29] with minor modifications. 2–5 flies per group were decapitated and placed into 1.5 ml tubes before adding 100 μl cold PBS. Samples were homogenized and 10 μl of supernatant were transferred to a new 1.5 ml tube for the protein level measurement. The remaining supernatant was heated at 70 °C for 10 min and then centrifuged at 14,000 rpm for 3 min at 4 °C. 60 μl of resulting supernatant was collected. Amyloglucosidase digestion solution was prepared by mixing 1.5 μl amyloglucosidase enzyme with 5 ml of 100 mM sodium citrate (pH = 5.0). 10 μl of collected supernatant of the glycogen sample was diluted in 90 μl of PBS in a separate 1.5 ml tube. 100 μl of amyloglucosidase solution was added and samples were incubated while shaking at 55 °C for 1 h. For quantification of glucose, 20 μl of remaining supernatant was added to a 96 well plate in duplicates and were diluted in 60 μl PBS. Similarly, 20 μl of incubated glycogen sample was added to the plate in duplicates and 100 μl prepared GO reagent (o-Dianisidine/Glucose Oxidase/Peroxidase, Sigma Aldrich, St. Louis, MO; GAGO20-1KT) was added to all samples. The plate was incubated at 37 °C for 30–60 min and 10 μl of 12 N sulfuric acid were added before absorbance was measured at 540 nm [29].
2.5. Photography
Age-matched control and knockdown flies were imaged using an AM Scope camera (AM Scope, MU1003).
2.6. Quantification of body weight
A group of five to ten age-matched flies was placed in a pre-weighed 1.5 ml tube. Body weight was calculated as mg per fly. Data was collected from 5 to 10 groups/genotype/sex.
2.7. Quantification of wing morphology
The frequency of unexpanded wings was determined by dividing the number of flies with abnormal wings by the total number of flies in the group. Two to five-day old flies were used.
2.8. Proboscis extension response (PER) assay
Two-day old flies raised on standard media were transferred to vials containing 0.7% agar after 24 h of fasting. A single fly was transferred by use of an oral aspirator and trapped in a 200 μl pipette tip with the head exposed. 1 μl of sucrose solution (150 mM) mixed with 3% erioglaucine (Sigma Aldrich, St. Louis, MO; 861146) was placed on the edge of the pipette tip within range of the proboscis. Proboscis extension and duration of feeding were observed for approximately 5 min.
2.9. Blue dye feeding assay
For each experiment, two-day old flies of 9–16 were fasted for 24 h on 0.7% agar. To measure baseline food intake without fasting, flies were kept on standard media and allowed to feed ad libitum. Feeding vials were prepared by placing a curled strip of filter paper or a pad (Figure 2A) in the center of an empty vial. 400 μl of the blue dye food solution (150 mM sucrose + 3% erioglaucine) was pipetted onto the filter paper. Flies were flipped into the vials and allowed to feed for 5 min if previously fasted, and 10 min if previously fed ad libitum. Flies were immediately incapacitated on dry ice and feeding frequency was determined by dividing the number of flies with blue bellies by the total number of flies. The flies were then individually placed in 1.5 ml tubes, homogenized in 50 μl of water, then centrifuged at 14,000 rpm for 5 min. To measure absorbance, 10 μl of supernatant was transferred to a 96 well plate and diluted in 40 μl of water. Absorbance was measured at 630 nm.
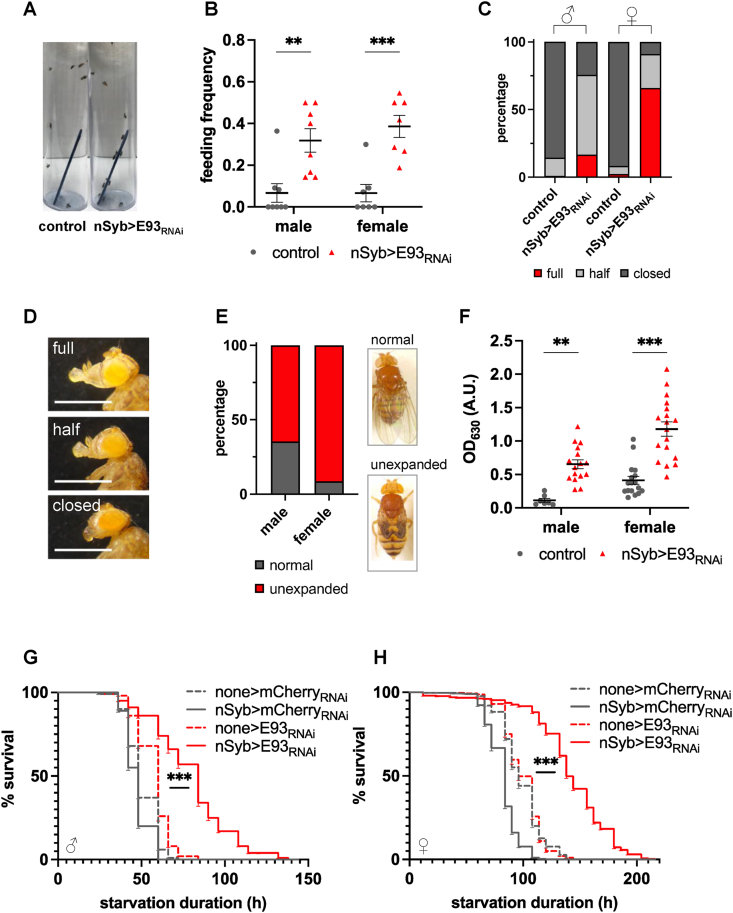
Figure 2.
Neuron-specific knockdown of E93 increases feeding frequency, food intake, and starvation survival. A–B:nSyb>E93RNAi flies are more attracted to food (A) and show a higher feeding frequency than the control (nSyb>mCherryRNAi) (B). Each data point represents one experiment of 9–16 flies. The bars indicate the mean ± S.E.M. C:nSyb>E93RNAi flies constantly feed as indirectly measured by % of flies with extended proboscis. N = 144–187 males, 83–100 females. D: Representative images of proboscises in each state of closed, half-open, and fully open. Scale bar = 100 μm. E: The % unexpanded wings in nSyb>E93RNAi flies. N = 82 males, 113 females. F:nSyb>E93RNAi flies have increased food intake. After 10 min of feeding, the flies were homogenized, and the food intake was measured by absorbance of the blue dye at 630 λ. none>E93RNAi flies were used as controls. Each data point represents one sample. The bars indicate the mean ± S.E.M. G–H: Both males (G) and females (H) of nSyb>E93RNAi survive starvation better than all three controls. B, F: ∗∗p < 0.005, ∗∗∗p < 0.001, ns = not significant by Student t-test. G, H: ∗∗∗p < 0.001 by log-rank test.
2.10. Quantification of proboscis extension
Flies were placed on dry ice to immediately capture their proboscises positions. The three positions of closed, half open, fully open were determined by the degree of proboscis extension.
2.11. Starvation assay
Starvation vials were prepared with 0.7% agar. Age-matched males and virgin females were collected and separately placed into starvation vials in 5 groups of 12 flies/vial for a total of 60 flies/sex/genotype. The number of deaths in each group was recorded three times a day until all flies were dead. The assay was performed on five separate cohorts of flies totaling 300 flies/sex/genotype.
2.12. Endurance and climbing speed assays
The Power Tower exercise platform was used to assess endurance [30,31]. Briefly, eight vials of 20 flies each were placed onto the Power Tower machine and were monitored every 10–15 min for exhaustion. A vial was defined as ‘exhausted’ once less than 20% of the flies in the vial (4 out of 20) were climbing above one centimeter from the top of their food. Each vial was graphed as a separate data point and the average time to exhaustion for a group was compared using a two-way ANOVA (to account for the effects of genotype and sex) with a post-hoc Tukey's multiple comparisons analysis in GraphPad Prism (GraphPad Software, San Diego, CA). The rapid negative geotaxis assay was used to evaluate climbing speed [30]. Briefly, cohorts of five vials with 20 flies each, were transferred to clear vials mounted in the RING apparatus. Flies were allowed 1 min for assimilation to the new environment. Then the vials were briskly tapped down (to move all flies to the bottom of their vials) and a picture was taken 2 s after the last tap. Six consecutive trials were conducted for each group, generating six pictures. The pictures were processed using ImageJ (U. S. National Institutes of Health, Bethesda, MD) to generate data representing climbing height, then analyzed using 2-way ANOVA with Tukey's multiple comparisons test.
2.13. Drosophila activity monitoring (DAM) assay and analyses
Flies were raised on standard media with 12 h light and 12 h dark condition. During the end of the light period, 2-day old flies were placed in the DAM system (TriKinetics Inc., Waltham, MA) where a single fly is loaded in each DAM tube with food on one end. Horizontal locomotor activity was measured by infra-red beam breaks in individual flies under conditions of 12 h light and 12 h dark or in constant darkness for 3 days. Because some nSyb>E93RNAi flies became unhealthy after being kept in the tube more than 5 days, we did not continuously record activity under light and dark condition followed by constant darkness. Naïve 2 days old flies were used for each lighting condition. The average number of beam breaks/6 min from 16 to 73 flies of 1–5 cohorts of each genotype and sex were used to generate actograms (ClockLab, Actimetrics, Wilmette, IL). Files that died during 3 days of recording were excluded from the group averaged actogram. Double plotted group averaged actograms were generated from 1 to 3 cohorts for each genotype and sex with 30 min bin and scaled plot (0–120 counts/30 min).
2.14. Fertility/receptivity assay
Ten pairs of males and females were placed in the presence of food in a vial and allowed to mate for 24 h. 5–7 days later, the fertility is determined by production of progeny. For female receptivity, 10 to 24 pairs of 2–4-day-old control males (nSyb>mCherryRNAi) and 2–3-day-old females (nSyb>mCherryRNAi or nSyb>E93RNAi) were placed in the presence of food in a vial or in a 12-well plate for 3 h. Then the parents were removed, and the progeny were counted.
2.15. Mating (courtship behavior) assay
The 2–5 days old males of each genotype were individually housed for 1–3 nights. The males and females were briefly anesthetized with CO2, then each male was paired with a control female. Each pair was placed into each well of a standard 6 well plate either in the presence or absence of food in the center. The flies were given 15–30 min to be fully recovered and then their mating behavior was recorded for 10 min. The number of mating behavior was manually scored when the male exhibited a sequence of two or more components of mating behavior: orienting toward the female, tracking the female, extending his wing and singing to the female, contacting the female, and mating [32,33]. This simple method allows us to monitor the mating behavior of E93RNAi males based on the behavioral components which do not involve wings.
2.16. Temperature sensitive GAL80ts experiments
The fly stock was generated by crossing the tubP-GAL80ts (#7019) to nSybGAL4 (#51635). The tubP-GAL80ts; nSybGAL4 were set up for mating in a bottle with either UAS-E93RNAi flies or UAS-mCherryRNAi flies either at 19 °C or at 30 °C. 3–5 days later the parents were removed, and the bottles were kept at the same temperature until wandering larvae appeared. For the constant 19 °C or 30 °C experiments, the bottles were kept at the same temperature and the F1s were collected. For temporal regulation of E93 RNAi at and after the third instar larvae (L3), once wandering larvae appeared, the bottles were moved from the initial temperature to the final temperature; either from 19 °C to 30 °C, or 30 °C to 19 °C. Then the bottles were kept under the new temperature until the flies eclosed. The F1s were collected within 4 h of post-eclosion and kept at the temperature for at least 3 days (72 h) before their body weight and circadian rhythm were measured. Only the F1s that eclosed in first two days at 30 °C and the F1s that eclosed in first three days at 19 °C which should have been L3s when the bottles were moved were used for the measurement of body weight and circadian rhythm.
For temporal regulation of E93 RNAi in adult stage, the flies raised at the initial temperature were collected within 3 h of eclosion then moved to the final temperature (either from 19 °C to 30 °C, or from 30 °C to 19 °C). The flies were kept at the final temperature for at least 3 days before their body weight and circadian rhythm were measured.
2.17. Statistical analysis
We examined whether all experimental groups were normally distributed using Sturges' class number and a symmetric distribution. All groups appeared to be normally distributed. F test was used to determine equality of variances for two independent groups. Comparisons between control and test groups were carried out using an unpaired two sample t-test with unequal variance. Log-rank test was used to validate the significance in difference in survivals (Figure 2G–H). Two-way ANOVA was used for Figure 1, Figure 4B. Analysis of endurance and speed (Figure 4) was conducted using a two-way ANOVA (to account for effects of genotype and sex) with post-hoc Tukey's multiple comparisons.
Figure 1.
Neuron-specific knockdown of E93 increases body weight and energy stores. A: Body weights of (3-day old) of males and females of nSyb>E93RNAi are increased compared to all RNAi only controls (none>E93RNAi and none>mCherryRNAi) and the GAL4 driver control (nSyb>mCherryRNAi). Each data point represents one group of 5 flies. The bars indicate the mean ± S.E.M. ∗∗∗p < 0.001 by two-way ANOVA. B: Representative images of males (♂) and females (♀) of control (nSyb>mCherryRNAi) and nSyb>E93RNAi flies. Scale bar = 1 mm. C–D: Time-course of body weight measurements in males (C) and females (D) showing a slight (male) or no (female) difference in initial body weights compared to controls (none>E93RNAi). Body weights gradually increase in nSyb>E93RNAi flies compared to controls. Errors are S.E.M. ∗∗∗p < 0.001 by Student t-test. E: Triglyceride (TAG) levels are increased in nSyb>E93RNAi males but not in nSyb>E93RNAi females when compared to controls (nSyb>mCherryRNAi). F: Glycogen levels are increased in nSyb>E93RNAi females but not in nSyb>E93RNAi males when compared to controls (nSyb>mCherryRNAi). E–F: Each data point represents one group of 5 males or one group of 2–5 females. The bars indicate the mean ± S.E.M. ∗p < 0.05, ∗∗p < 0.005, ns = not significant by Student t-test.
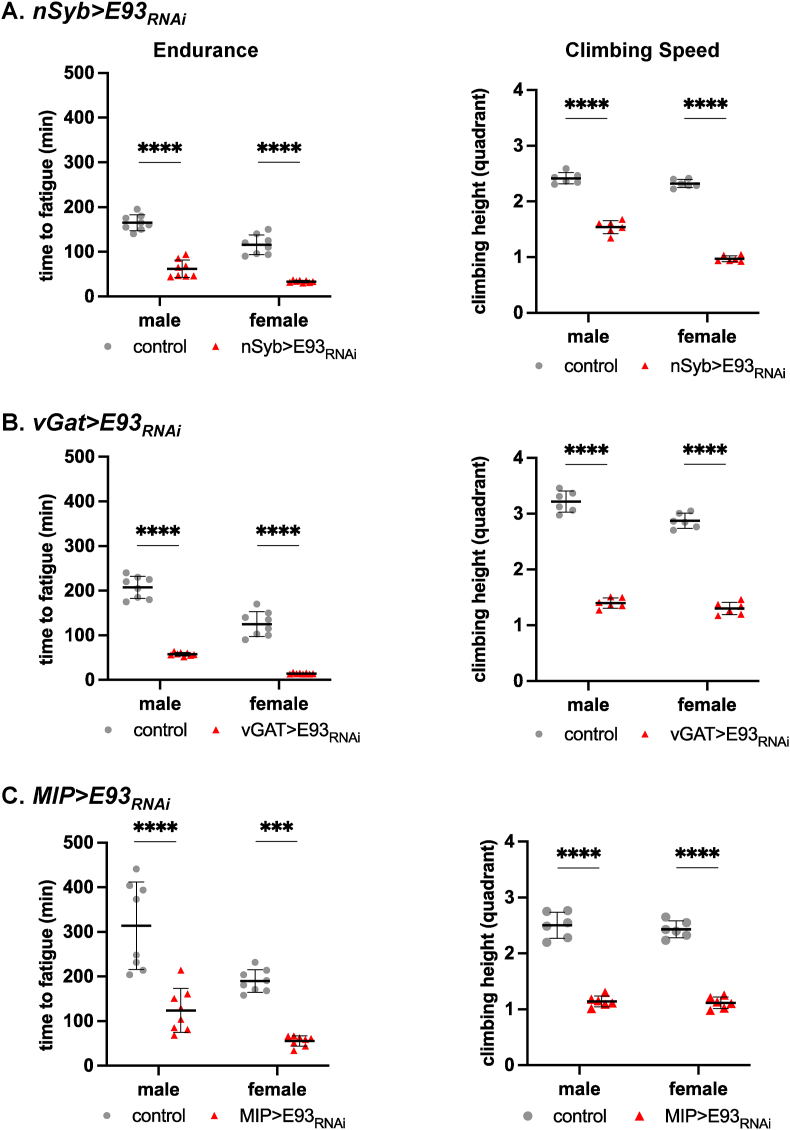
Figure 4.
Reduced E93 expression in neurons decreases endurance and climbing speed in adult flies. A: Pan-neuronal knockdown of E93 significantly reduced endurance and climbing speed in nSyb>E93RNAi male and female flies. B: GABAergic neuron-specific knockdown of E93 significantly reduced endurance and climbing speed in vGAT>E93RNAi male and female flies. C: MIP neuron-specific knockdown of E93 significantly reduced endurance and climbing speed in MIP>E93RNAi male and female flies. All controls were generated by crossing the genetic background of the GAL4 line with E93RNAi (none>E93RNAi). ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, by two-way ANOVA with post-hoc Tukey's multiple comparisons test.
3. Results
3.1. A starvation-resistance screen of selected Drosophila orthologs enriched in the rodent ventromedial hypothalamus (VMH) identified E93
To identify new metabolic regulators, we performed an RNAi-based screen of 70 Drosophila genes orthologous to the genes enriched in ventromedial hypothalamus (VMH) in mice [34,35] (GSE96627) (Table S1). The fly orthologs were determined based on the previous reports [36]. We focused on VMH because of its essential roles in metabolic homeostasis. Using the GAL4-UAS system, we crossed the line carrying UAS (Upstream Activating Sequence) flanked with an RNAi construct of each gene to a GAL4 line to reduce the expression. To limit the reduction of the gene to the nervous system, we crossed the UAS-RNAi line of each gene to the nSyb-GAL4 driver and reduced the expression specifically in the neurons. To validate the screen, we used multiple controls: Adipokinetic hormone (Akh), the functional homolog of glucagon in fly whose mutation results in resistance to starvation [37], as a positive control, sh, a homolog of Kcna1 enriched in VMH whose mutation results in reduced lifespan, as a negative control [38]. UAS-GFP and UAS-mCherryRNAi were used as controls for UAS and for RNAi, respectively. Neuronal knockdown of upd1, the fly leptin, and NPF were used as controls for metabolism.
We screened the F1 progeny of the cross between nSyb-GAL4 and the UAS-RNAi of each gene for their survival for long-term starvation on the premise that coping with this extreme metabolic challenge requires the fittest metabolic regulation. Therefore, this screen would allow us to identify genes that contribute to any aspect of metabolic homeostasis by controlling metabolic rates, feeding, reproduction and other related traits. Importantly, the underlying mechanisms and genetic programs for metabolic homeostasis (e.g., insulin signaling) are highly conserved between invertebrates and mammals [[39], [40], [41]].
Reduced expression of upd1 enhanced starvation survival in both males and females (Figure S1). In addition, GPR155/LYCHOS, an ortholog of anchor that was a strong positive hit in the screen, was recently discovered to play an important role in cholesterol signaling to metabolism [42]. These results support that our screen platform was well-tuned to identify a new metabolic regulator. The screen identified E93 as the strongest hit next to the positive control (Figure S1).
3.2. Neuron-specific reduction of E93 expression increases body weight and energy stores
When we reduced the expression of E93 pan-neuronally using an nSyb-GAL4 driver and a UAS-E93 RNAi (from here, nSyb>E93RNAi), the flies became obese; their body weight was increased compared to the three controls of UAS, GAL4, and genetic background (Figure 1A). Because all three controls show very similar body weights, we used either the Berlin K strain carrying UAS-E93 RNAi without any GAL4 driver (none>E93RNAi) or nSyb>mCherryRNAi as a control depending on the experiment. nSyb>mCherryRNAi, nSyb>E93RNAi and none>E93RNAi lines were outcrossed to w Berlin for five generations to isogenize the background.
Three days after eclosion, both male and female nSyb>E93RNAi flies were visibly obese (Figure 1B). The body weight of nSyb>E93RNAi flies gradually increased each day after eclosion (Figure 1C–D). When we measured abdomen width, both knockdown males and females had increased abdomen size (Figure S2A). Compared to controls, female knockdowns had significantly enlarged abdomens beginning one day after eclosion, but both nSyb>E93RNAi and controls retained similar numbers of mature eggs (14.4 ± 5.3, N = 18, vs 14.9 ± 4.3, N = 13, Supplementary Methods), indicating that the enlarged abdomen observed in knockdowns was not due to an increased number of retained eggs.
When we measured the levels of triacylglycerides (TAG) and glycogen, we found that males had more TAG, whereas females had more glycogen than controls (Figure 1E–F). These results suggest that neuronal E93 regulates energy stores and that the energy stores may be regulated in a sexually dimorphic manner.
3.3. Neural specific knockdown of E93 increases food intake
Next, we asked whether the increased body weight and energy stores in nSyb>E93RNAi flies were due to an increase in food intake. When nSyb>E93RNAi flies were placed in a vial with a pad containing sucrose solution (150 mM) and blue dye, a higher percentage of nSyb>E93RNAi flies gathered to the pad than controls (Figure 2A). Consistent with this observation, nSyb>E93RNAi flies showed increased feeding frequency measured by the percentage of total fed flies with blue abdomens (Figure 2B). Furthermore, the majority of nSyb>E93RNAi flies stayed on the pad throughout the experiment, whereas the control flies quickly ate and left within several seconds (Supplementary Video 1). The proboscis, the feeding organ, of nSyb>E93RNAi flies was fully extended at a higher frequency in nSyb>E93RNAi flies than in the control, suggesting that E93 knockdown flies are feeding more frequently than the controls (Figure 2C–D).
The Capillary Feeding (CAFE) assay is commonly used to assess food intake. It relies on the ability of flies to reach a suspended food source and hang upside down long enough to feed until satiated [43]. As nSyb>E93RNAi flies have unexpanded wings (Figure 2E), we were unable to perform the CAFE assay reliably. Instead, we modified a blue dye feeding assay and determined the amount of individual food intake by measuring dye absorbance at λ630 (see Methods) [44]. Virgin male and female nSyb>E93RNAi flies fasted for 24 h had increased food intake compared to controls (Figure 2F).
We used the proboscis extension response (PER) assay to further examine the hyperphagia of nSyb>E93RNAi flies in detail. Flies were individually trapped in a pipet tip and presented with 1 μl of sucrose solution containing blue dye under the proboscis [26]. The duration of proboscis extension and the volume of consumed food were measured over 5 min by subtracting the remaining volume from the 1 μl originally presented. Although the number of samples we tested with this method was small, we were able to observe a striking difference between nSyb>E93RNAi flies and controls. All nSyb>E93RNAi flies maintained extended proboscises for the entire 5 min (100%, 5 out of 5) and 80% (4 out of 5) of flies finished 1 μl within 5 min. On the other hand, only 40% (2 out of 5) of the control flies had a single bout of feeding lasting 30 s or less and consumed less than 0.4 μl (Supplementary Videos 2–3). The remaining 60% did not extend their proboscis to feed at all.
In females, changes in midgut size are often associated with increased food intake when nutritional demand is high [45]. The average diameter of the midgut of nSyb>E93RNAi females was increased compared to controls, suggesting an increase in food intake (Figure S2B–C). In males increased food intake is often associated with increased defecation rate [46]. nSyb>E93RNAi males show an increased defecation rate compared to controls, suggestive of increased food intake (Figure S2D). As observed in the screen, both male and female nSyb>E93RNAi flies survived starvation longer than all the controls (Figure 2G–H), suggesting that their increased energy stores rendered them starvation resistant. Taken together, our data indicate that neuronal E93 is required to control food intake and energy stores.
3.4. E93 acts mainly in GABA-ergic and MIP-producing neurons
To determine the sites of E93 action, we screened 17 GAL4 lines by which the expression of the E93 RNAi construct was limited to subsets of specific classes of neurons and/or endocrine cells. We used glycogen levels in females as a readout, reasoning that the big difference in glycogen levels between nSyb>E93RNAi and the control would provide us a reliable resolution for detection. The 17 lines include the three classical neurotransmitter-producing neurons: vGlut (glutamate), ChAT (acetylcholine) and vGAT (GABA); three monoamine-producing neurons: ple (dopamine), Tdc2 (octopamine/tyramine) and Trh (serotonin); and the mushroom body specific line, OK107, which is important for learning and memory. The other ten drivers were selected based on their known roles in feeding and metabolism, and include cells producing MIP (myoinhibitory peptide) [26], Akh (Adipokinetic hormone) [47], Eth (Ecdysone triggering hormone) [48], Phm (ecdysone producing cells of the prothoracic glands) [49], Tk (Tachykinin) [50,51], NPF (neuropeptide F) [52], Fmrfa (FMRFamid neuropeptide), Dilp2 (Insulin-like peptide), Lk (Leucokinin), and SIFaR (SIFamide receptor) [53]. To control for the effect of different GAL4s, we used UAS-mCherryRNAi as a control for each driver. Of note, UAS-mCherryRNAi (none>mCherryRNAi) flies had comparable metabolic parameters, including body weight and starvation response, to other controls (Figure 1, Figure 2G–H).
Flies with a knockdown of E93 in GABA-ergic neurons or in MIP neurons had increased glycogen levels (Figure S3A). Specific GABA-ergic interneurons are known to play a critical role in inhibiting feeding [22]. Indeed, body weight and feeding frequency were increased in both male and female vGAT>E93RNAi flies compared to controls (Figure 3A–C). In addition, a higher percentage of vGAT>E93RNAi males and females extended their proboscises than controls (93% of vGAT>E93RNAi (N = 57) vs. 2% of vGAT>mCherryRNAi (N = 46)). The abdomen size of the vGAT>E93RNAi females increased without an increase of the number of retained eggs (Figure S3B–C). These results validate the use of glycogen levels as a readout to identify the sites of E93 action in regulation of food intake and body weight. Similar to nSyb>E93RNAi flies, vGAT>E93RNAi flies also exhibited a wing phenotype; 50% of males and 82% of females had unexpanded wings while 100% of controls had normal wings (N = 32 for males, N = 50 for females). These similarities between nSyb>E93RNAi flies and vGAT>E93RNAi flies in increased body weight, attraction to food, and unexpanded wings indicate that GABA-ergic neurons are one of the main sites of E93 action.
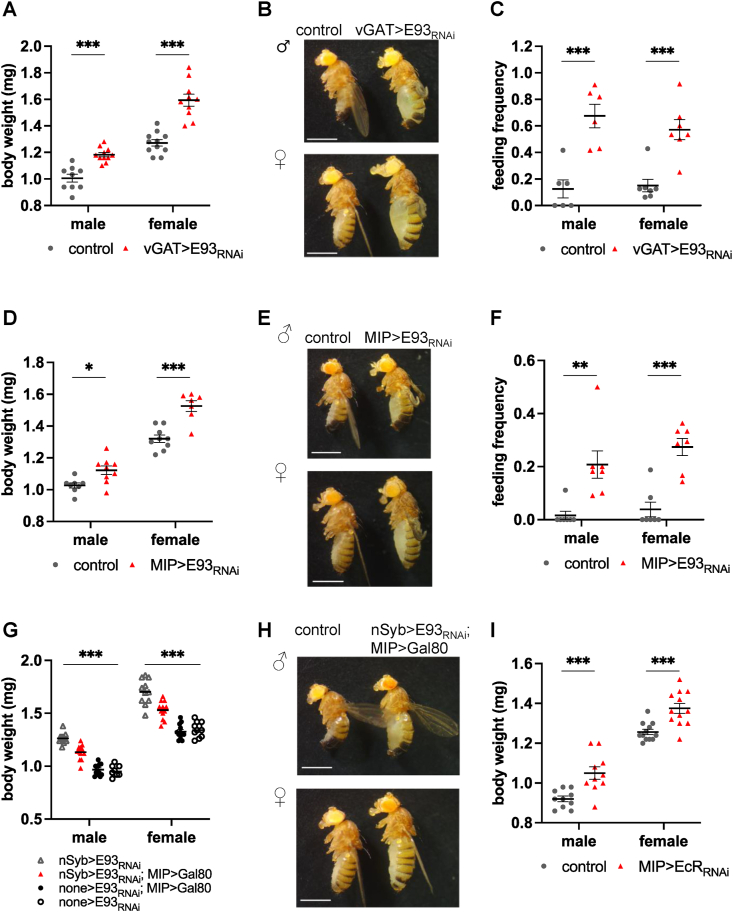
Figure 3.
Loss of E93 in GABA-ergicand MIP neurons underliesmetabolic changes. A–C:vGAT>E93RNAi flies phenocopies nSyb>E93RNAi flies with increased body weight (A), obese appearance (B), and increased feeding frequency (C). vGAT>mCherryRNAi was used as controls. A: Each data point represents one group of 5 flies. C: Each data point represents one experiment of 9–16 flies. The bars indicate the mean ± S.E.M. B: Scale bar = 1 mm. D–F:MIP>E93RNAi phenocopies nSyb>E93RNAi with increased body weight (D), obese appearance (E), and increased feeding frequency (F). MIP>mCherryRNAi flies were used as controls. D: Each data point represents one group of 5 flies. F: Each data point represents one experiment of 9–16 flies. The bars indicate the mean ± S.E.M. ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.001 by Student t-test. G–H:MIP>GAL80 partially but significantly reverses the increased body weight phenotype of nSyb>E93RNAi. Each data point represents one group of 5 flies. The bars indicate the mean ± S.E.M. ∗∗p < 0.005, ∗∗∗p < 0.001 by Two-way ANOVA. H: Scale bar = 1 mm. I:MIP>EcRRNAi phenocopies MIP>E93RNAi with increased body weight. MIP>mCherryRNAi flies were used as controls. Each data point represents one group of 5 flies. The bars indicate the mean ± S.E.M. ∗∗∗p < 0.001 by Student t-test.
Next, we examined flies with E93 knocked down in MIP neurons. MIP/allatostatin-B homologs are present in almost all animals [54], and serve versatile roles related to metabolism such as feeding and metamorphosis [55]. In Drosophila, MIP is expressed in approximately 70 neurons in the areas important for food sensation such as the antennal lobe and subesophageal zone (SEZ), and regulates mating receptivity, satiety, fat storage, and sleep [[26], [27], [28]]. Both males and females of MIP>E93RNAi are obese and feed more (Figure 3D–F), indicating MIP neurons are also where E93 acts. The MIP>E93RNAi females resemble the previously reported phenotypes of the MIP mutant females [26]. In addition, MIP>E93RNAi females have increased abdomen size without changing in the number of retained mature eggs (Figure S3D–E).
To confirm that MIP neurons are a site of E93 action, we introduced GAL80, a repressor of GAL4, only in MIP neurons in nSyb>E93RNAi flies. MIP-enhancer-driven GAL80 represses GAL4 driven by nSyb in MIP neurons and thus expression of E93 is restored only in MIP neurons, while nSyb-GAL4 continues to drive E93 RNAi in all other neurons. Introducing GAL80 only in MIP neurons partially but significantly reverses the body weight phenotype of nSyb>E93RNAi flies (Figure 3G–H), confirming that MIP neurons are one of the sites of E93 action.
3.5. Steroid nuclear receptor controls metabolism in MIP neurons
The steroid hormone, ecdysone, binds to the ecdysone receptor (EcR) to initiate metamorphosis and induce expression of E93 [11]. Ecdysone and EcR regulate many essential functions not only during metamorphosis but also in adult flies. Ecdysone is secreted mainly from the fat body in males and ovaries in females, and controls reproduction and metabolism [[56], [57], [58]]. To test whether neuronal EcR regulates metabolic homeostasis potentially regulating E93, we reduced expression of EcR in GABA-ergic neurons and MIP neurons. GABA-ergic neuron-specific knockdown of EcR (vGAT>EcRRNAi) produces lethality, thus we could not investigate the role of EcR in feeding and metabolism in these cells. However, the MIP neuron-specific knockdown of EcR (MIP>EcRRNAi) partly phenocopied MIP>E93RNAi showing increased body weight (Figure 3I, Figure S4A), suggesting a genetic interaction of E93 with EcR in MIP neurons in regulation of metabolism. Similar to nSyb>EcRRNAi females, MIP>EcRRNAi females had increased abdomen size without changes in the number of retained mature eggs (Figure S4B–C). However, we found that the feeding frequency of MIP>EcRRNAi was either not different from the controls (Figure S4D, female) or slightly reduced (Figure S4D, male). The mechanisms underlying this dissociation between weight gain and food intake are unknown. One possibility is that MIP>EcRRNAi flies have altered patterns of food intake and activity (see below), which might have obscured the correct timing for us to measure food intake. Alternatively, as EcR plays a significant role from the birth, knockdown of EcR in MIP neurons throughout development could result in adaptations or defects that mask the feeding phenotype seen in adult MIP>E93RNAi. It is also possible that the increased food intake seen in MIP>E93RNAi might be independent to EcR.
GABA is important for controlling food intake [22,23] and is secreted from some MIP neurons [59]. We therefore examined whether GABA produced in MIP neurons contributes to the observed nSyb>E93RNAi phenotypes. However, no differences in body weight were observed in flies with knockdown of the vesicular transporter of GABA (vGAT) in MIP neurons, suggesting that GABA released by MIP neurons might not contribute to the obesity phenotype of MIP>E93RNAi flies (Figure S4E–F).
3.6. Reduced E93 expression in neurons decreases adult endurance and climbing speed
Studies suggest that metabolic dysregulation can be associated with impaired activity such as climbing [60,61]. We also showed that exercise training improves adult physiology and therefore provides a useful assessment of systemic adaptation of an adult [30]. The obesity phenotype with increased body weight and energy stores led us to ask whether neuronal E93 was required to maintain wild-type endurance and speed in induced and involuntary assessments of exercise performance in adults. When we tested male and female nSyb>E93RNAi flies, using an established paradigm to assess exercise performance [30,31], they showed dramatically reduced endurance and speed (Figure 4A). To determine whether E93 is required for exercise performance in the same GABA-ergic and MIP neurons where it is required for obesity and feeding phenotypes, we next tested vGAT>E93RNAi and MIP>E93RNAi flies. Both males and females of vGAT>E93RNAi (Figure 4B) or MIP>E93RNAi (Figure 4C) reduced endurance and speed to a similar extent as nSyb>E93RNAi flies (Figure 4A), indicating that E93 expression in these neurons is critical for adult exercise performance.
3.7. Reduced expression of E93 in neurons alters locomotor activity and disrupts circadian rhythm
The reciprocal interaction between biological rhythm and metabolism is conserved in many animals and metabolic abnormality is often associated with abnormal locomotive activity [60,62]. For example, Clock mutant mice are obese [63] and high-fat diet disrupts the activity and feeding rhythms [64,65]. Studies in Drosophila also reveal evolutionarily conserved mechanisms of circadian rhythm controlled by metabolism [[66], [67], [68], [69]]. When we measured the daytime and nighttime activity of nSyb>E93RNAi flies using the Drosophila Activity Monitoring (DAM) system, both females and males showed overall reduced activity under 12 h light and 12 h dark condition compared to the controls (Figure 5A). Similar reductions in activity were observed in MIP>E93RNAi flies (Figure 5C). However, no obvious change in activity was observed in vGAT>E93RNAi flies and only a modest reduction in activity was observed in MIP>EcRRNAi flies (Figure 5B and D). Interestingly, the nighttime activity of vGAT>E93RNAi was elevated (Figure 5B). GABA activity is required to promote sleep by suppressing wake-promoting neurons such as pigment dispersing factor (PDF) producing neurons and monoaminergic neurons such as dopamine or octopamine [25,70]. Therefore, the increased nighttime activity observed in vGAT>E93RNAi suggests that E93 could play a role in neuronal GABA activity to control food intake as well as to promote sleep.
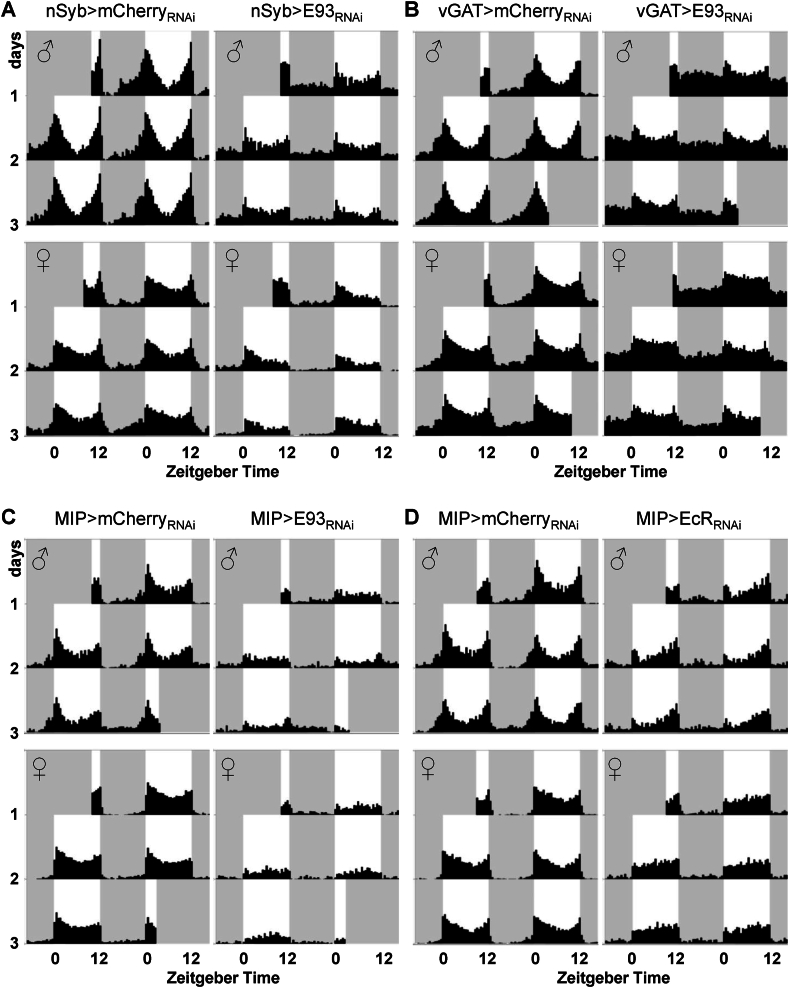
Figure 5.
Reduced E93 expression in neurons attenuates morning and evening circadian anticipation. Beam breaks were recorded every 6 min from individual fly using the DAM assay system under 12 h light and 12 h dark conditions. Group average double-plotted actograms were generated with 30 min bins and plotted scale 0–120 counts/30 min. The x-axis indicates Zeitgeber time and y-axis indicates days. Control (mCherry RNAi; left) and E93 or EcR knockdown (right) flies are plotted by sex (males (♂): top, females (♀): bottom). White background indicates light phase and gray background indicates dark phase. A: Pan-neuronal knockdown of E93 expression using the nSyb-GAL4 driver. nSyb>mCherryRNAi males (N = 27, 2 cohorts), nSyb>mCherryRNAi females (N = 29, 2 cohorts), nSyb>E93RNAi males (N = 31, 2 cohorts), and nSyb>E93RNAi females (N = 31, 2 cohorts). B: Knockdown of E93 expression in GABA-ergic neurons. vGAT>mCherryRNAi males (N = 39, 3 cohorts), vGAT>mCherryRNAi females (N = 73, 5 cohorts), vGAT>E93RNAi males (N = 45, 3 cohorts), and vGAT>E93RNAi females (N = 64, 5 cohorts). C: Knockdown of E93 expression in MIP neurons. MIP>mCherryRNAi males (N = 31, 2 cohorts), MIP>mCherryRNAi females (N = 32 2 cohorts), MIP>E93RNAi males (N = 32, 2 cohorts), and MIP>E93RNAi females (N = 32, 2 cohorts). D: Knockdown of EcR expression in MIP neurons. MIP>mCherryRNAi males (N = 32, 2 cohorts), MIP>mCherryRNAi females (N = 28, 2 cohorts), MIP>EcRRNAi males (N = 30, 2 cohorts), and MIP>EcRRNAi females (N = 32, 2 cohorts).
Under standard 12 h light and 12 h dark cycles, Drosophila melanogaster expresses a bimodal activity pattern with increased anticipatory activity prior to lights-on (dawn) and lights-off (dusk). Anticipations of dawn and dusk are controlled by different subsets of circadian pacemaker neurons, morning oscillator and evening oscillator, respectively [[71], [72], [73]]. While control flies exhibit stereotypical anticipatory activities, these behaviors are almost absent in nSyb>E93RNAi flies (Figure 5A), suggesting that autonomous circadian clocks in E93 knockdown flies could be disrupted. In fact, in constant darkness, the majority of nSyb>E93RNAi flies show nearly arrhythmic activity patterns (Figure 6A), and only a few flies exhibit low amplitude of rhythms (1 out of 16 knockdown males, 2 out of 15 knockdown females, Figure S5). Similar to nSyb>E93RNAi flies, vGAT>E93RNAi, MIP>E93RNAi, MIP>EcRRNAi flies show moderate anticipation for lights-on or off (Figure 5B–D), suggesting that their circadian rhythms are also disrupted. vGAT>E93RNAi, MIP>E93RNAi, MIP>EcRRNAi flies also exhibited damped circadian rhythms under constant darkness compared to that of controls (Figure 6B–D). While nSyb>E93RNAi, vGAT>E93RNAi, MIP>E93RNAi, MIP>EcRRNAi flies all show disrupted circadian rhythm, the disruptions in nSyb>E93RNAi flies under constant darkness are the most severe.
Figure 6.
Reduced E93 expression in neurons disrupts circadian rhythm. Group average double-plotted actograms of flies in constant darkness are shown. Flies raised in 12 h light and 12 h dark condition were placed in DAM system during the light phase and released in constant darkness at the time of lights off. The white box indicates the last light cycle before releasing flies in constant darkness. A: Pan-neuronal knockdown of E93 expression using the nSyb-GAL4 driver. nSyb>mCherryRNAi males (N = 45, 3 cohorts), nSyb>mCherryRNAi females (N = 46, 3 cohorts), nSyb>E93RNAi males (N = 59, 4 cohorts), and nSyb>E93RNAi females (N = 61, 4 cohorts). B: Knockdown of E93 expression in GABA-ergic neurons. vGAT>mCherryRNAi males (N = 26, 2 cohorts), vGAT>mCherryRNAi females (N = 39, 3 cohorts), vGAT>E93RNAi males (N = 31, 2 cohorts), and vGAT>E93RNAi females (N = 44, 3 cohorts). C: Knockdown of E93 expression in MIP neurons. MIP>mCherryRNAi males (N = 31, 2 cohorts), MIP>mCherryRNAi females (N = 31 2 cohorts), MIP>E93RNAi males (N = 32, 2 cohorts), and MIP>E93RNAi females (N = 31, 2 cohorts). D: Knockdown of EcR expression in MIP neurons. MIP>mCherryRNAi males (N = 31, 2 cohorts), MIP>mCherryRNAi females (N = 31, 2 cohorts), MIP>EcRRNAi males (N = 30, 2 cohorts), and MIP>EcRRNAi females (N = 32, 2 cohorts). Other conditions are the same as the actograms in Figure 5.
Together, our results suggest that E93 in the two subsets of neurons, MIP-producing and GABA-ergic neurons, play an essential role in the regulation of metabolism, locomotive activity, and circadian rhythm all of which are essential for adult fitness.
3.8. E93 action is critical during L3 to adult transition
To determine the timing of E93 action, we used a GAL80ts line with a tubulin promoter (tubP) expressing GAL80ts ubiquitously [74]. GAL80ts is an inhibitor of GAL4, whose activity can be manipulated by temperature. At a permissive temperature (19 °C), GAL80ts is functional and thus inhibits nSyb-GAL4 activity in neurons. This blocks neuronal E93 RNAi and thus E93 functions normally. At a nonpermissive temperature (30 °C), however, GAL80ts is no longer functional resulting in nSyb-GAL4 activation and E93 RNAi in neurons. To pinpoint the timing of E93 action, we shifted temperature either at a late larval stage (the third instar larvae L3 and wandering larvae) when E93 expression is known to be highest [20] or within 4 h after eclosion as an adult. The GAL80ts; nSyb-GAL4>E93RNAi flies raised at constant 19 °C showed no difference in body weight compared to the control, GAL80ts; nSyb-GAL4>mCherryRNAi, (Figure 7A), whereas GAL80ts;nSyb-GAL4>E93RNAi flies raised at constant 30 °C became obese (Figure 7B). These results confirmed that the obesity is indeed caused by reduction of E93 expression. The phenotypes did not change when we shifted the temperature in adults; the obese flies remained obese, and the normal flies remained normal (Figure 7C–D). When we shifted the temperature at the late larva stage, however, GAL80ts;nSyb-GAL4>E93RNAi flies became obese (Figure 7E). Furthermore, if we raised the flies at 30 °C until L3 then shifted to 19 °C, the flies did not become obese, indicating that the transition beginning from late L3 is the critical period of E93 action (Figure 7F).
Figure 7.
E93 function is critical during larval-to-adult transition. A: The body weights are not different between tubP-GAL80ts; nSybGAL4>E93RNAi flies (shown as GAL80ts:nSyb>E93RNAi) and tubP-GAL80ts; nSybGAL4>mCherryRNAi (shown as GAL80ts:nSyb>mCherryRNAi) flies when both were raised at constant 19 °C, a permissive temperature, throughout life. B:tubP-GAL80ts; nSybGAL4>E93RNAi flies are heavier than tubP-GAL80ts; nSybGAL4>mCherryRNAi flies when both were raised at constant 30 °C, a non-permissive temperature, throughout life. C: The body weights are not different between tubP-GAL80ts; nSybGAL4>E93RNAi flies and tubP-GAL80ts; nSybGAL4>mCherryRNAi flies when both were raised at 19 °C and then moved to 30 °C immediately after eclosion. D:tubP-GAL80ts; nSybGAL4>E93RNAi flies are heavier than tubP-GAL80ts; nSybGAL4>E93RNAi flies when both were raised at 30 °C and then moved to 19 °C immediately after eclosion. E:tubP-GAL80ts; nSybGAL4>E93RNAi flies are heavier than tubP-GAL80ts; nSybGAL4>mCherryRNAi flies when both were raised at 19 °C and then moved to 30 °C at the L3 stage. F: The body weights are not different between tubP-GAL80ts; nSybGAL4>E93RNAi flies and tubP-GAL80ts; nSybGAL4>mCherryRNAi flies when both were raised at 30 °C and then moved to 19 °C at the L3 stage. A–F: Each data point represents one group of 7–10 flies. Flies were 3–8 days old. The bars indicate the mean ± S.E.M. ns: not significant, ∗∗∗p < 0.001 by Student t-test.
Next, we tested whether the obesity phenotype correlates with other physiological defects such as circadian rhythm. When we examined circadian rhythm of the GAL80ts;nSyb-GAL4>E93RNAi flies raised at constant 19 °C or 30 °C, under the constant darkness conditions, the activity pattern correlated with the obesity phenotype; the flies raised at 19 °C showed a similar activity pattern to that of the control, whereas the flies raised at 30 °C show disrupted activity pattern (Figure S6A–B). Consistent with that result, the flies raised at 19 °C then moved to 30 °C at L3 stage showed a disrupted activity pattern (Figure S6C), whereas the flies raised at 30 °C then moved to 19 °C at L3 stage did not (Figure S6D). These results indicate that E93 action at the transition is critical for normal body weight and circadian activity.
3.9. Reduced expression of E93 in neurons reduces reproductive activity
The neuronal knockdown of E93 results in such a dramatic and systemic failure in the expression of adult metabolism and physiology that we hypothesize that the brain of nSyb>E93RNAi flies might remain in the larval state focusing on growth and eating instead of on adult behavior such as mating. As shown in Figure 7, E93 critically functions for regulation of metabolism during larval-to-adult transition when the larval brain develops into the adult brain, supporting this hypothesis. When we examined the courtship behavior in the presence of food [33], the control males exhibited multiple courtship components and sequence within 10-minute recording span, such as following the female, touching her, performing wing dance, and mounting [32] (Figure 8A, Supplementary Video 4, right). However, nSyb>E93RNAi males did not interact with the female but often went for food (Figure 8A, Supplementary Video 4, left).
Figure 8.
Reduced E93 expression in neurons reduces reproductive behavior. A–C: The number of exhibition of mating behavior of males in the presence of food (see the Methods and Supplementary videos). A:nSyb>mCherryRNAi males (control) and nSyb>E93RNAi, N = 12 for both. 3–5 days old. B:vGAT>mCherryRNAi males (control) N = 9, and vGAT>E93RNAi males N = 8. 2–3 days old. C:MIP>mCherryRNAi males (control) and MIP>E93RNAi. N = 7 for both. 3 days old. Each data point represents a single male's behavior. D–F: The number of exhibition of mating behavior in the absence of food. D: nSyb>mCherryRNAi (control) and nSyb>E93RNAi, N = 9 for both. 2–3 days old. E: vGAT>mCherryRNAi (control) N = 26, and vGAT>E93RNAi N = 28. 3–5 days old. F: MIP>mCherryRNAi (control) and MIP>E93RNAi N = 16 for both. 3 days old. The bars indicate the mean ± S.E.M. ∗∗p < 0.01, ∗∗∗∗p < 0.0001 by Student t-test. G: Model A larva and an adult have distinct goals of growth and reproduction, respectively. This difference in goals directs the behavior and activity of each form to maximize fitness. Larvae's main activity is feeding to increase the mass, whereas the adults' is coordinated behavior such as circadian rhythmicity, optimum fitness, and energy stores for mating success. Our study suggests that E93 in GABA-ergic neurons and MIP-producing neurons acts as a switch critical for this transition.
Because nSyb>E93RNAi flies show increased attraction to food (Figure 2A), We next tested whether the lack of courtship behaviors in nSyb>E93RNAi males was due to a preference for food over mates. When we performed the mating assay in the absence of food, however, nSyb>E93RNAi males still did not show courtship behavior (Figure 8D, Supplementary Video 5). Instead, they usually rested in the corner, suggesting that the lack of courtship behavior in nSyb>E93RNAi males is not due to a preference for food over a mate.
The lack of courtship behavior resulted in failure of reproduction. When a female and male pair was placed in a vial with food for 24 h, the control pairs produced progeny, as did the nSyb>E93RNAi female with a control male. However, nSyb>E93RNAi males with a control or a nSyb>E93RNAi female failed to produce progeny (Figure S7A). Because wings play a significant role in courtship behavior and because nSyb>E93RNAi flies do not have functional wings, we tested whether the infertility of nSyb>E93RNAi males was due to their lack of functional wings. When we removed wings of control males, however, the de-winged controls produced progeny, indicating that in our setup, the wings are dispensable for mating (Figure S7B). Both vGAT>E93RNAi males and MIP>E93RNAi males fail to exhibit mating behavior regardless of food (Figure 8B–C, E–F, Supplementary Videos 6–9), suggesting these two neuron populations could also be the main action sites of E93 to regulate reproductive activity.
During mating, a female fly evaluates the male partner via several pre-mating responses such as flapping, escaping, and kicking, before she eventually accepts the male [75,76]. Although nSyb>E93RNAi females could reproduce (Figure S7A), it is possible that their receptivity is reduced. We tested the receptivity indirectly by a modified fertility assay, where a pair of flies was given only 3 h for mating. We reason that if the receptivity of nSyb>E93RNAi females were reduced, the mating rate would be also reduced. In two independent experiments where 20 or 24 females of nSyb>mCherryRNAi or nSyb>E93RNAi were each individually paired with a nSyb>mCherryRNAi male for 3 h, 15 and 20 nSyb>mCherryRNAi females produced progeny (15/20, 20/24, an average of 79% mating rate). However, only 2 nSyb>E93RNAi females produced progeny (2/20, 2/24, an average of 10% mating rate, Figure S7C)). This result suggests that reduction of neuronal E93 expression reduces reproductive activity both in males and females.
4. Discussion
In this study, we report E93 as a new monogenic factor of metabolic syndrome, identified from a targeted screen of 70 selected genes orthologous to mammalian genes enriched in VMH. Neuron-specific knockdown of E93 causes a systemic failure of metabolic homeostasis, leading to phenotypes such as hyperphagia, increased energy stores, and impaired exercise endurance. Neuronal E93 is also required for circadian rhythmicity, and for mating behavior for males, indicating E93 is critical for the animals to exhibit adult behaviors. Screening of 17 different GAL4 drivers revealed that E93 function in metabolism, adult physiology and behavior is primarily via its action in GABA-ergic and MIP-producing neurons, both of which have been implicated in regulation of feeding, metabolism, and sleep [22,24,26,28,70]. Consistent with its known role in metamorphosis, E93 action during the 3rd instar (L3) and metamorphosis is the critical developmental time window for its normal body weight. E93 partly interacts with the ecdysone receptor (EcR) in MIP neurons, linking steroid signal to a peptidergic signal. Increased frequency of feeding and the resulting increased food intake most likely underlies the obesity in flies with neuron-specific knockdowns of E93.
Neuron-specific knockdown of E93 also exhibits multiple physiological defects such as impaired exercise endurance, reduced climbing speed, disrupts circadian rhythm, and reduced reproduction activity. Similar defects in exercise endurance, climbing speed, circadian rhythm, and reproduction activity were observed in flies with GABA-specific or MIP-specific knockdowns of E93, supporting the idea that these neurons are the main sites of E93 action. Disruption of circadian rhythm is most pronounced when the flies are exposed to constant darkness (Figure 6). All nSyb>E93RNAi, vGAT>E93RNAi, MIP>E93RNAi, and MIP>EcRRNAi flies failed to show lights-on and off anticipatory behavior but exhibited either arrhythmicity or low amplitude rhythms under constant darkness. It is possible that reduced activity itself underlies the circadian phenotype. For instance, in a DAM assay, flies that stay near the food and spend a significantly longer time feeding than the controls would show decreased numbers of beam crosses. However, vGAT>E93RNAi flies showed disrupted circadian rhythms, yet their activity levels were comparable to controls, suggesting that the arrhythmicity in males or the low amplitudes in females in constant darkness could be independent of overall activity. Taken together, our data from vGAT>E93RNAi and nSyb>E93RNAi flies suggest that as least some part of disrupted circadian rhythm of nSyb>E93RNAi is likely associated with their global metabolic abnormalities rather than overall reduction of activity.
A mammalian homolog of E93 is LCoR (Ligand-dependent Corepressor) and has been shown to directly interact with nuclear receptors critical for metabolism [[77], [78], [79], [80]]. This suggests that E93 might regulate circadian rhythm by interacting with the key nuclear receptors required for the metabolic clock [81,82]. Indeed, the fly orthologs of the key nuclear receptors, E75 (Retinoic acid-related Orphan Receptors in mammals) and DHR51 (Drosophila Hormone Receptor 51, REV-ERB in mammals) have conserved roles in regulating circadian rhythm [83,84].
Although, the detailed neuro-molecular mechanisms by which E93 regulates these pleiotropic metabolic phenotypes require further investigation, based on its role in body weight regulation from the temperature sensitive experiment, we suggest that E93 is the key switch in shaping the adult brain to confer the adult metabolism. As a transcription factor, E93 could directly control gene expression in coordination with specific nuclear receptors and switch the neuron's profile from larva to adult, as suggested in the model (Figure 8G). Our result that obesity can be induced by reducing neuronal E93 expression only during L3 and metamorphosis also supports this hypothesis. Indeed, a recent study shows that several larva-enriched genes are abnormally upregulated during metamorphosis in the E93 deletion mutant [17]. Interestingly, many of these genes are also implicated in neural fate determination (e.g. squeeze, ko), metabolism (e.g. secreted decoy of Inr), and circadian rhythm (e.g. clock), supporting a role for E93 in the transition from larval to adult metabolism and physiology. One way by which E93 could control the transition from the larval brain to the adult brain is to establish the adult neuronal circuitry by controlling neurite pruning as its C. elegans ortholog does [13]. In fact, the nervous system undergoes dramatic rewiring via steroid hormones during adolescence in many animals to switch the focus from growth to reproduction [85,86]. Drosophila also undergoes intensive remodeling of the nervous system during metamorphosis a stage in which E93 plays its essential roles [87].
Together, our results provide insights into how a transcription factor acting in specific sets of neurons in the nervous system shapes and regulates adult metabolism, physiology, and behavior.
Funding
This work was supported by the National Institutes of Health (NIH) through R01DK100659 (JE), P30DK127984 (JE), R01NS114527 (SY), R01AA019526 (AR), R01AA026818 (AR), R01AG059683 (RW), 2T32HL120822 (TC), and by the National Science Foundation (NSF) through IOS-1931115 (SY).
Ethics statement
No animal work in this study requires specific regulations.
CRediT authorship contribution statement
Cecilia Yip: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Conceptualization. Steven C. Wyler: Writing – review & editing, Supervision, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Katrina Liang: Investigation. Shin Yamazaki: Writing – review & editing, Writing – original draft, Validation, Software, Funding acquisition, Formal analysis. Tyler Cobb: Investigation, Funding acquisition, Formal analysis. Maryam Safdar: Writing – original draft, Investigation, Formal analysis. Aarav Metai: Investigation, Formal analysis. Warda Merchant: Methodology, Investigation. Robert Wessells: Writing – review & editing, Writing – original draft, Supervision, Funding acquisition, Formal analysis, Conceptualization. Adrian Rothenfluh: Writing – review & editing, Supervision, Funding acquisition. Syann Lee: Writing – review & editing, Supervision, Conceptualization. Joel Elmquist: Supervision, Funding acquisition, Conceptualization. Young-Jai You: Writing – review & editing, Writing – original draft, Validation, Supervision, Resources, Methodology, Investigation, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgement
We thank Drs. Kim J, Fujikawa T, and Sieber M, and the Sieber lab members for invaluable discussions, Drs. Smith D, Buszczak M, and Kramer H, Ms. Chelsea Limboy for technical help. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. We used FlyBase to obtain information on multiple genes including E93. We thank Korea Drosophila Resource Center, and Dr. Kim Y (GIST) for fly strains.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2024.101939.
Contributor Information
Joel Elmquist, Email: Joel.Elmquist@UTSouthwestern.edu.
Young-Jai You, Email: Young-Jai.You@UTsouthwestern.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- 1.Tennessen J.M., Baker K.D., Lam G., Evans J., Thummel C.S. The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metab. 2011;13(2):139–148. doi: 10.1016/j.cmet.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tennessen J.M., Bertagnolli N.M., Evans J., Sieber M.H., Cox J., Thummel C.S. Coordinated metabolic transitions during Drosophila embryogenesis and the onset of aerobic glycolysis. G3 (Bethesda) 2014;4(5):839–850. doi: 10.1534/g3.114.010652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H., Rai M., Buddika K., Sterrett M.C., Luhur A., Mahmoudzadeh N.H., et al. Lactate dehydrogenase and glycerol-3-phosphate dehydrogenase cooperatively regulate growth and carbohydrate metabolism during Drosophila melanogaster larval development. Development. 2019;146(17) doi: 10.1242/dev.175315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Covell C.V. Peterson Field Guides: Eastern Moths. Houghton Mifflin Company; 1984. p. 49. [Google Scholar]
- 5.Tennessen J.M., Thummel C.S. Coordinating growth and maturation - insights from Drosophila. Curr Biol. 2011;21(18):R750–R757. doi: 10.1016/j.cub.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jindra M., Palli S.R., Riddiford L.M. The juvenile hormone signaling pathway in insect development. Annu Rev Entomol. 2013;58:181–204. doi: 10.1146/annurev-ento-120811-153700. [DOI] [PubMed] [Google Scholar]
- 7.Riddiford L.M., Truman J.W., Mirth C.K., Shen Y.C. A role for juvenile hormone in the prepupal development of Drosophila melanogaster. Development. 2010;137(7):1117–1126. doi: 10.1242/dev.037218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Texada M.J., Koyama T., Rewitz K. Regulation of body size and growth control. Genetics. 2020;216(2):269–313. doi: 10.1534/genetics.120.303095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elmquist J.K., Elias C.F., Saper C.B. From lesions to leptin: hypothalamic control of food intake and body weight. Neuron. 1999;22(2):221–232. doi: 10.1016/s0896-6273(00)81084-3. [DOI] [PubMed] [Google Scholar]
- 10.Siegmund T., Lehmann M. The Drosophila Pipsqueak protein defines a new family of helix-turn-helix DNA-binding proteins. Dev Genes Evol. 2002;212(3):152–157. doi: 10.1007/s00427-002-0219-2. [DOI] [PubMed] [Google Scholar]
- 11.Baehrecke E.H., Thummel C.S. The Drosophila E93 gene from the 93F early puff displays stage- and tissue-specific regulation by 20-hydroxyecdysone. Dev Biol. 1995;171(1):85–97. doi: 10.1006/dbio.1995.1262. [DOI] [PubMed] [Google Scholar]
- 12.Urena E., Manjon C., Franch-Marro X., Martin D. Transcription factor E93 specifies adult metamorphosis in hemimetabolous and holometabolous insects. Proc Natl Acad Sci U S A. 2014;111(19):7024–7029. doi: 10.1073/pnas.1401478111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kage E., Hayashi Y., Takeuchi H., Hirotsu T., Kunitomo H., Inoue T., et al. MBR-1, a novel helix-turn-helix transcription factor, is required for pruning excessive neurites in Caenorhabditis elegans. Curr Biol. 2005;15(17):1554–1559. doi: 10.1016/j.cub.2005.07.057. [DOI] [PubMed] [Google Scholar]
- 14.Conway E., Jerman E., Healy E., Ito S., Holoch D., Oliviero G., et al. A family of vertebrate-specific polycombs encoded by the LCOR/LCORL genes balance PRC2 subtype activities. Mol Cell. 2018;70(3):408–421 e408. doi: 10.1016/j.molcel.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Plassais J., Kim J., Davis B.W., Karyadi D.M., Hogan A.N., Harris A.C., et al. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat Commun. 2019;10(1):1489. doi: 10.1038/s41467-019-09373-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song Y., Shan S., Zhang Y., Liu W., Ding W., Ren W., et al. Ligand-dependent corepressor acts as a novel corepressor of thyroid hormone receptor and represses hepatic lipogenesis in mice. J Hepatol. 2012;56(1):248–254. doi: 10.1016/j.jhep.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Lam G., Nam H.J., Velentzas P.D., Baehrecke E.H., Thummel C.S. Drosophila E93 promotes adult development and suppresses larval responses to ecdysone during metamorphosis. Dev Biol. 2021;481:104–115. doi: 10.1016/j.ydbio.2021.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nystrom S.L., Niederhuber M.J., McKay D.J. Expression of E93 provides an instructive cue to control dynamic enhancer activity and chromatin accessibility during development. Development. 2020;147(6) doi: 10.1242/dev.181909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pahl M.C., Doyle S.E., Siegrist S.E. E93 integrates neuroblast intrinsic state with developmental time to terminate MB neurogenesis via autophagy. Curr Biol. 2019;29(5):750–762 e753. doi: 10.1016/j.cub.2019.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frise E., Hammonds A.S., Celniker S.E. Systematic image-driven analysis of the spatial Drosophila embryonic expression landscape. Mol Syst Biol. 2010;6:345. doi: 10.1038/msb.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jafari S., Alkhori L., Schleiffer A., Brochtrup A., Hummel T., Alenius M. Combinatorial activation and repression by seven transcription factors specify Drosophila odorant receptor expression. PLoS Biol. 2012;10(3) doi: 10.1371/journal.pbio.1001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pool A.H., Kvello P., Mann K., Cheung S.K., Gordon M.D., Wang L., et al. Four GABAergic interneurons impose feeding restraint in Drosophila. Neuron. 2014;83(1):164–177. doi: 10.1016/j.neuron.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung S.K., Scott K. GABAA receptor-expressing neurons promote consumption in Drosophila melanogaster. PLoS One. 2017;12(3) doi: 10.1371/journal.pone.0175177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agosto J., Choi J.C., Parisky K.M., Stilwell G., Rosbash M., Griffith L.C. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci. 2008;11(3):354–359. doi: 10.1038/nn2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parisky K.M., Agosto J., Pulver S.R., Shang Y., Kuklin E., Hodge J.J., et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60(4):672–682. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Min S., Chae H.S., Jang Y.H., Choi S., Lee S., Jeong Y.T., et al. Identification of a peptidergic pathway critical to satiety responses in Drosophila. Curr Biol. 2016;26(6):814–820. doi: 10.1016/j.cub.2016.01.029. [DOI] [PubMed] [Google Scholar]
- 27.Jang Y.H., Chae H.S., Kim Y.J. Female-specific myoinhibitory peptide neurons regulate mating receptivity in Drosophila melanogaster. Nat Commun. 2017;8(1):1630. doi: 10.1038/s41467-017-01794-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh Y., Yoon S.E., Zhang Q., Chae H.S., Daubnerova I., Shafer O.T., et al. A homeostatic sleep-stabilizing pathway in Drosophila composed of the sex peptide receptor and its ligand, the myoinhibitory peptide. PLoS Biol. 2014;12(10) doi: 10.1371/journal.pbio.1001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tennessen J.M., Barry W.E., Cox J., Thummel C.S. Methods for studying metabolism in Drosophila. Methods. 2014;68(1):105–115. doi: 10.1016/j.ymeth.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Damschroder D., Cobb T., Sujkowski A., Wessells R. Drosophila endurance training and assessment of its effects on systemic adaptations. Bio Protoc. 2018;8(19) doi: 10.21769/BioProtoc.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piazza N., Gosangi B., Devilla S., Arking R., Wessells R. Exercise-training in young Drosophila melanogaster reduces age-related decline in mobility and cardiac performance. PLoS One. 2009;4(6) doi: 10.1371/journal.pone.0005886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall J.C. The mating of a fly. Science. 1994;264(5166):1702–1714. doi: 10.1126/science.8209251. [DOI] [PubMed] [Google Scholar]
- 33.Lin H.H., Kuang M.C., Hossain I., Xuan Y., Beebe L., Shepherd A.K., et al. A nutrient-specific gut hormone arbitrates between courtship and feeding. Nature. 2022;602(7898):632–638. doi: 10.1038/s41586-022-04408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S., Bookout A.L., Lee C.E., Gautron L., Harper M.J., Elias C.F., et al. Laser-capture microdissection and transcriptional profiling of the dorsomedial nucleus of the hypothalamus. J Comp Neurol. 2012;520(16):3617–3632. doi: 10.1002/cne.23116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segal J.P., Stallings N.R., Lee C.E., Zhao L., Socci N., Viale A., et al. Use of laser-capture microdissection for the identification of marker genes for the ventromedial hypothalamic nucleus. J Neurosci. 2005;25(16):4181–4188. doi: 10.1523/JNEUROSCI.0158-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N., et al. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee G., Park J.H. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167(1):311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trout W.E., Kaplan W.D. A relation between longevity, metabolic rate, and activity in shaker mutants of Drosophila melanogaster. Exp Gerontol. 1970;5(1):83–92. doi: 10.1016/0531-5565(70)90033-1. [DOI] [PubMed] [Google Scholar]
- 39.Guarente L., Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408(6809):255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 40.Clancy D.J., Gems D., Harshman L.G., Oldham S., Stocker H., Hafen E., et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292(5514):104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 41.Wullschleger S., Loewith R., Hall M.N. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 42.Shin H.R., Citron Y.R., Wang L., Tribouillard L., Goul C.S., Stipp R., et al. Lysosomal GPCR-like protein LYCHOS signals cholesterol sufficiency to mTORC1. Science. 2022;377(6612):1290–1298. doi: 10.1126/science.abg6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ja W.W., Carvalho G.B., Mak E.M., de la Rosa N.N., Fang A.Y., Liong J.C., et al. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci U S A. 2007;104(20):8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hadjieconomou D., King G., Gaspar P., Mineo A., Blackie L., Ameku T., et al. Enteric neurons increase maternal food intake during reproduction. Nature. 2020;587(7834):455–459. doi: 10.1038/s41586-020-2866-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White M.A., Bonfini A., Wolfner M.F., Buchon N. Drosophila melanogaster sex peptide regulates mated female midgut morphology and physiology. Proc Natl Acad Sci U S A. 2021;118(1) doi: 10.1073/pnas.2018112118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dus M., Lai J.S., Gunapala K.M., Min S., Tayler T.D., Hergarden A.C., et al. Nutrient sensor in the brain directs the action of the brain-gut axis in Drosophila. Neuron. 2015;87(1):139–151. doi: 10.1016/j.neuron.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galikova M., Diesner M., Klepsatel P., Hehlert P., Xu Y., Bickmeyer I., et al. Energy homeostasis control in Drosophila adipokinetic hormone mutants. Genetics. 2015;201(2):665–683. doi: 10.1534/genetics.115.178897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mena W., Diegelmann S., Wegener C., Ewer J. Stereotyped responses of Drosophila peptidergic neuronal ensemble depend on downstream neuromodulators. Elife. 2016;5 doi: 10.7554/eLife.19686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taghert P.H., Hewes R.S., Park J.H., O'Brien M.A., Han M., Peck M.E. Multiple amidated neuropeptides are required for normal circadian locomotor rhythms in Drosophila. J Neurosci. 2001;21(17):6673–6686. doi: 10.1523/JNEUROSCI.21-17-06673.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song W., Veenstra J.A., Perrimon N. Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 2014;9(1):40–47. doi: 10.1016/j.celrep.2014.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qi W., Wang G., Wang L. A novel satiety sensor detects circulating glucose and suppresses food consumption via insulin-producing cells in Drosophila. Cell Res. 2021;31(5):580–588. doi: 10.1038/s41422-020-00449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung B.Y., Ro J., Hutter S.A., Miller K.M., Guduguntla L.S., Kondo S., et al. Drosophila neuropeptide F signaling independently regulates feeding and sleep-wake behavior. Cell Rep. 2017;19(12):2441–2450. doi: 10.1016/j.celrep.2017.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nassel D.R., Pauls D., Huetteroth W. Neuropeptides in modulation of Drosophila behavior: how to get a grip on their pleiotropic actions. Curr Opin Insect Sci. 2019;36:1–8. doi: 10.1016/j.cois.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 54.Williams E.A. Function and distribution of the Wamide neuropeptide superfamily in metazoans. Front Endocrinol (Lausanne) 2020;11:344. doi: 10.3389/fendo.2020.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Audsley N., Weaver R.J. Neuropeptides associated with the regulation of feeding in insects. Gen Comp Endocrinol. 2009;162(1):93–104. doi: 10.1016/j.ygcen.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 56.Belles X., Piulachs M.D. Ecdysone signalling and ovarian development in insects: from stem cells to ovarian follicle formation. Biochim Biophys Acta. 2015;1849(2):181–186. doi: 10.1016/j.bbagrm.2014.05.025. [DOI] [PubMed] [Google Scholar]
- 57.Buszczak M., Freeman M.R., Carlson J.R., Bender M., Cooley L., Segraves W.A. Ecdysone response genes govern egg chamber development during mid-oogenesis in Drosophila. Development. 1999;126(20):4581–4589. doi: 10.1242/dev.126.20.4581. [DOI] [PubMed] [Google Scholar]
- 58.Sieber M.H., Spradling A.C. Steroid signaling establishes a female metabolic state and regulates SREBP to control oocyte lipid accumulation. Curr Biol. 2015;25(8):993–1004. doi: 10.1016/j.cub.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Croset V., Treiber C.D., Waddell S. Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. Elife. 2018;7 doi: 10.7554/eLife.34550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y., Hoffmann J., Li Y., Stephano F., Bruchhaus I., Fink C., et al. Octopamine controls starvation resistance, life span and metabolic traits in Drosophila. Sci Rep. 2016;6 doi: 10.1038/srep35359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo S., Zhang S., Zhuang Y., Xie F., Wang R., Kong X., et al. Muscle PARP1 inhibition extends lifespan through AMPKalpha PARylation and activation in Drosophila. Proc Natl Acad Sci U S A. 2023;120(13) doi: 10.1073/pnas.2213857120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ladabaum U., Mannalithara A., Myer P.A., Singh G. Obesity, abdominal obesity, physical activity, and caloric intake in US adults: 1988 to 2010. Am J Med. 2014;127(8):717–727 e712. doi: 10.1016/j.amjmed.2014.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turek F.W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308(5724):1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kohsaka A., Laposky A.D., Ramsey K.M., Estrada C., Joshu C., Kobayashi Y., et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6(5):414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 65.Pendergast J.S., Branecky K.L., Yang W., Ellacott K.L., Niswender K.D., Yamazaki S. High-fat diet acutely affects circadian organisation and eating behavior. Eur J Neurosci. 2013;37(8):1350–1356. doi: 10.1111/ejn.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bass J., Takahashi J.S. Circadian integration of metabolism and energetics. Science. 2010;330(6009):1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Konopka R.J., Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68(9):2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dubowy C., Sehgal A. Circadian rhythms and sleep in Drosophila melanogaster. Genetics. 2017;205(4):1373–1397. doi: 10.1534/genetics.115.185157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Allada R., Emery P., Takahashi J.S., Rosbash M. Stopping time: the genetics of fly and mouse circadian clocks. Annu Rev Neurosci. 2001;24:1091–1119. doi: 10.1146/annurev.neuro.24.1.1091. [DOI] [PubMed] [Google Scholar]
- 70.Ni J.D., Gurav A.S., Liu W., Ogunmowo T.H., Hackbart H., Elsheikh A., et al. Differential regulation of the Drosophila sleep homeostat by circadian and arousal inputs. Elife. 2019;8 doi: 10.7554/eLife.40487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stoleru D., Peng Y., Nawathean P., Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438(7065):238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- 72.Stoleru D., Peng Y., Agosto J., Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431(7010):862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- 73.Yoshii T., Rieger D., Helfrich-Forster C. Two clocks in the brain: an update of the morning and evening oscillator model in Drosophila. Prog Brain Res. 2012;199:59–82. doi: 10.1016/B978-0-444-59427-3.00027-7. [DOI] [PubMed] [Google Scholar]
- 74.McGuire S.E., Mao Z., Davis R.L. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE. 2004;2004(220) doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- 75.Connolly K., Cook R. Rejection responses by female drosophila-melanogaster - their ontogeny, causality and effects upon behavior of courting male. Behaviour. 1973;44(1–2):142–166. [Google Scholar]
- 76.Ishimoto H., Kamikouchi A. Molecular and neural mechanisms regulating sexual motivation of virgin female Drosophila. Cell Mol Life Sci. 2021;78(10):4805–4819. doi: 10.1007/s00018-021-03820-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cao H., Zhang S., Shan S., Sun C., Li Y., Wang H., et al. Ligand-dependent corepressor (LCoR) represses the transcription factor C/EBPbeta during early adipocyte differentiation. J Biol Chem. 2017;292(46):18973–18987. doi: 10.1074/jbc.M117.793984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shalom-Barak T., Liersemann J., Memari B., Flechner L., Devor C.E., Bernardo T.M., et al. Ligand-dependent corepressor (LCoR) is a rexinoid-inhibited peroxisome proliferator-activated receptor gamma-retinoid X receptor alpha coactivator. Mol Cell Biol. 2018;38(9) doi: 10.1128/MCB.00107-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Asim M., Hafeez B.B., Siddiqui I.A., Gerlach C., Patz M., Mukhtar H., et al. Ligand-dependent corepressor acts as a novel androgen receptor corepressor, inhibits prostate cancer growth, and is functionally inactivated by the Src protein kinase. J Biol Chem. 2011;286(43):37108–37117. doi: 10.1074/jbc.M111.292771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fernandes I., Bastien Y., Wai T., Nygard K., Lin R., Cormier O., et al. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol Cell. 2003;11(1):139–150. doi: 10.1016/s1097-2765(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 81.Sato T.K., Panda S., Miraglia L.J., Reyes T.M., Rudic R.D., McNamara P., et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43(4):527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 82.Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 83.Kumar S., Chen D., Jang C., Nall A., Zheng X., Sehgal A. An ecdysone-responsive nuclear receptor regulates circadian rhythms in Drosophila. Nat Commun. 2014;5:5697. doi: 10.1038/ncomms6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Beuchle D., Jaumouille E., Nagoshi E. The nuclear receptor unfulfilled is required for free-running clocks in Drosophila pacemaker neurons. Curr Biol. 2012;22(13):1221–1227. doi: 10.1016/j.cub.2012.04.052. [DOI] [PubMed] [Google Scholar]
- 85.Sisk C.L., Foster D.L. The neural basis of puberty and adolescence. Nat Neurosci. 2004;7(10):1040–1047. doi: 10.1038/nn1326. [DOI] [PubMed] [Google Scholar]
- 86.Sisk C.L., Zehr J.L. Pubertal hormones organize the adolescent brain and behavior. Front Neuroendocrinol. 2005;26(3–4):163–174. doi: 10.1016/j.yfrne.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 87.Tissot M., Stocker R.F. Metamorphosis in drosophila and other insects: the fate of neurons throughout the stages. Prog Neurobiol. 2000;62(1):89–111. doi: 10.1016/s0301-0082(99)00069-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.