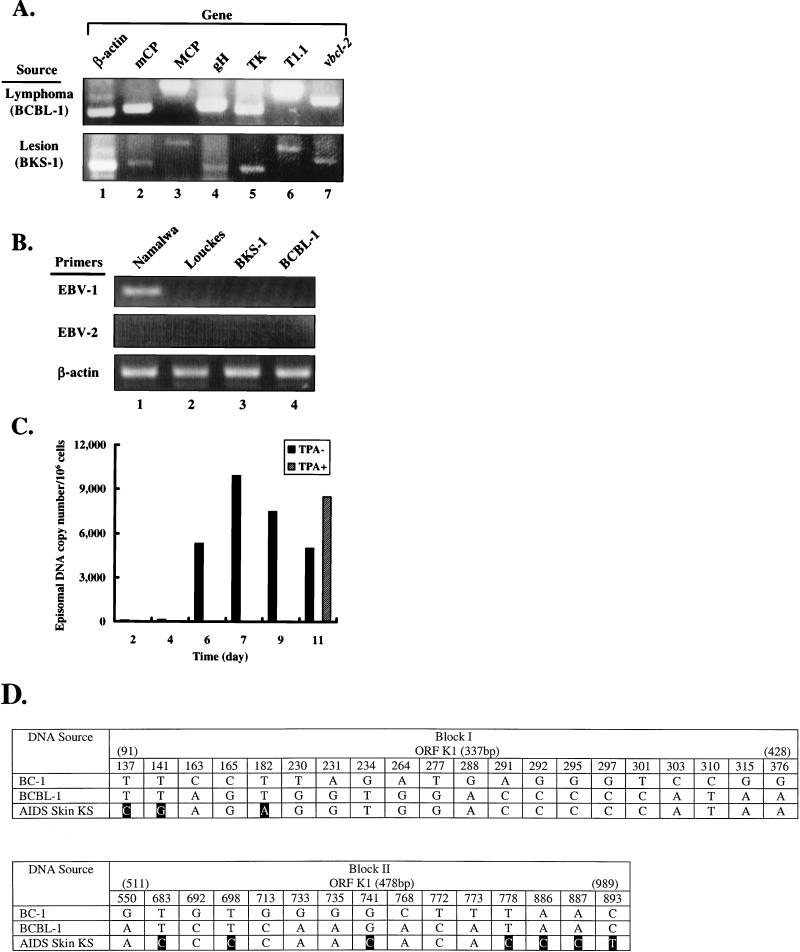
FIG. 1.
Transmission of KSHV DNA sequences from primary KS lesions into Louckes cells. PCR analysis was performed with 1 μg of total genomic DNA extracted from BCBL-1 and Louckes cells following cocultivation with primary KS lesions (BKS-1) as described in the text. PCR products were separated by electrophoresis on an ethidium bromide-stained agarose gel (2%). Numerous functional and structural KSHV genes were amplified from both cell lines (A), whereas EBV DNA was undetectable (B). EBV standard was generated from DNA isolated from Namalwa cells which contain two integrated EBV genomes per cell, and cellular β-actin was used as a control for PCRs. (C) Quantitative PCR analysis of KSHV DNA sequences was performed with the TaqMan PCR detection system (Applied Biosystems) according to the manufacturer’s instructions. Total genomic DNA was extracted from BKS-1 cells (106) at the indicated times following cocultivation, and PCR analyses were performed in triplicate, using a custom fluorescent probe and primers for ORF26 (mCP) of KSHV. At day 9, cells were cultured in fresh medium containing or lacking TPA (20 ng/ml) for 48 h prior to DNA isolation. (D) Sequence variability of ORF K1 among KSHV DNA samples from different sources. In this summary of the results of PCR sequencing analysis of two segments of the ORF K1 obtained from a cutaneous KS lesion and BCBL-1, only nucleotides present at variable positions are given, with positions that differ from the prototype KSHV sequence (GeneBank accession no. 1102887) enclosed in black boxes. Primers used and sizes of the PCR products are as follows: 5′-GTACAATCAAGATGTTCCTGTATG-3′ (sense) and 5′-ACAAGTGACTGTGTTTGATGG-3′ (antisense), giving a 337-bp product; and 5′-CCGTGTCACAAACTAAATACT-3′ (sense) and 5′-TATCTTACCTGAATGTCAGTACCA-3′ (antisense), giving a 478-bp product.