FIG. 1.
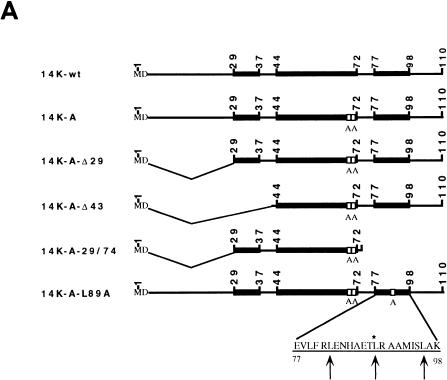
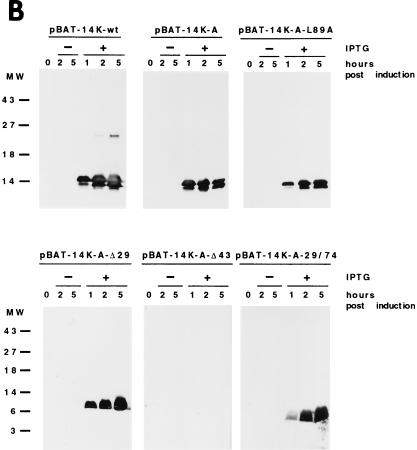
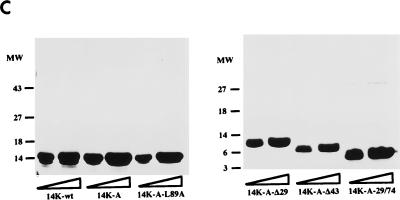
(A) Diagram representing mutant forms of the VV 14-kDa protein. Black bars indicate regions predicted as being α-helices in the protein secondary structure. Open boxes correspond to the point mutations introduced in the sequences, and the amino acids changed are indicated below the boxes. The three hepta repeat leucine residues present in the putative third α-helix are indicated by arrows, and the amino acid that was changed is indicated by the asterisk. (B) Expression in E. coli of mutant forms of the VV 14-kDa protein. E. coli BL21(DE3) cells, transformed with different plasmids, were grown to an optical density of 0.4 at 560 nm in the absence or presence of inducer IPTG (1 mM). At various times postinduction (0, 1, 2, and 5 h), aliquots were removed and centrifuged, cells were lysed, and proteins were fractionated by SDS-PAGE and visualized by immunoperoxidase staining after reaction with the 14-kDa-specific MAbC3. Wild-type and mutant forms of the 14-kDa protein were fractionated on SDS–13% PAGE gels, while deletion mutants were fractionated on SDS–15% PAGE gels. The mutant with a 43-amino-acid deletion at the N terminus does not show reactivity with the antibody in spite of high-level expression as revealed by Coomassie staining (not shown). Molecular weight (MW) markers are shown to the left. Plasmid origins are shown on top (see panel A). (C) Coomassie blue staining of purified 14-kDa proteins. The proteins were purified as described in Materials and Methods. Results for two different concentrations (10 and 20 μg/lane) of each protein are shown. The full-length proteins (wild type and point mutation mutants) were fractionated on SDS–13% PAGE gels, and mutant forms with deletions were fractionated on SDS–15% PAGE gels. Molecular weight (MW) markers are shown to the left.