Abstract
Background and Objectives:
Klebsiella pneumoniae is an opportunistic pathogen responsible for causing nosocomial and community-acquired infections. Its pathogenicity is associated with a variety of virulence factors and antibiotic resistance. The aim of the present study was to compare virulence attributes between ESBL and non-ESBL producing isolates.
Materials and Methods:
A total of 113 K. pneumoniae including 56 ESBL and 57 non ESBL-producers were collected in Bushehr province, Iran, from November 2017 to February 2019. Enzymatic profile, hypermucoviscosity and biofilm formation were investigated phenotypically. In addition, the presence of rmpA, aerobactin, kfu, allS, mrkD, ybtS, entB, iutA, fimH, wabG, wcaG, K1 and K2 genes were detected by PCR and sequencing.
Results:
There was no statistically significant difference in enzymatic profile between ESBL and non-ESBL producers. The prevalence of the hypermocoviscosity was lower among ESBL compared to non-ESBL producers but the intensity of biofilm was higher in the ESBL producers. Among the virulence genes, K1, rmpA, iutA, and aero were observed only in non-ESBLs. Moreover, the carriage of allS, K, K2, rmpA, iutA and aero genes was higher in hypermucoviscous in comparison with non hypermucoviscous isolates.
Conclusion:
The identification of potentially pathogenic isolates plays an important role in preventing their spread as well as the success of their treatment.
Keywords: Klebsiella pneumoniae, Biofilm, Extended-spectrum beta-lactamase, Virulence
INTRODUCTION
Klebsiella pneumoniae is one of the most important pathogens responsible for many community-acquired and healthcare-associated infections including pneumonia, urinary tract infection, abdominal infection, surgical wounds, liver abscesses and bacteremia (1). The presence of medical devices such as respiratory support equipment and various types of catheters greatly increases the risk of K. pneumoniae infections (2). K. pneumoniae pathogenicity is caused due to various virulence factors overcoming the patient’s innate immune system, hence, rendering the infection persistent. These factors include capsule, lipopolysaccharide, adhesins, siderophores and biofilm formation (3). The clinical manifestations and severity of the disease caused by K. pneumoniae are related to the number of expressed virulence factors (4). Along with the capsular polysaccharide which protects these organisms against the bactericidal effect of serum killing by professional phagocytes, the fimH-1 and mrkD genes, encoding type 1 and type 3 fimbrial adhesins play an important role via biofilm formation in the establishment of infection. Several other virulence-associated genes including those encoding regulators of mucoid phenotype A (rmpA), mucoviscosity-associated gene A (magA) located within an operon specific to serotype K1 capsule gene clusters, enterobactin biosynthesis gene (entB), yersiniabactin biosynthesis gene (ybtS), iron siderophores aerobactin gene (aero), an iron uptake gene (kfu), allantoin metabolism associated gene (allS) highly contributed to the pathogenicity and severity level of K. pneumoniae infections.
In parallel, over the past decade the prevalence of extended-spectrum β-lactamase (ESBL)-producing K. pneumoniae has been drastically increasing and thus causing a worldwide public health concern. Compared to planktonic bacteria, biofilm-forming bacteria have certain advantages (9) including protection against the host’s immune system and antimicrobial agents (2, 10). Additionally, in the biofilm environment the transfer of genetic material, especially by conjugative transfer, occurs easily due to the proximity of the bacteria. This, along with increasing the survival of the bacteria in the biofilm, might be an important factor in the widespread distribution of multi-resistant plasmids harboring ESBLs genes (9).
The simultaneous study of virulence and antimicrobial resistance, especially to third-generation cephalosporins, which are widely used in the treatment of K. pneumoniae infections (11), might provide better understanding of the relationship between virulence and β-lactam resistance. In addition, pathogenic microbes are able to cause damage to host tissues and establishment of infection through the secretion of various enzymes such as proteinases, lecithinases, gelatinase, lipases, and hemolysin (12). Therefore, the aims of the present study were to investigate the production of extracellular enzymes, biofilm production and virulence genes and compare virulence attributes between ESBL and non-ESBL producing K. pneumoniae isolates.
MATERIALS AND METHODS
Bacterial isolation and identification. This project was approved by the Ethical Committee of Bushehr University of Medical Sciences with reference number IR.BPUMS.REC.1398.136. A total of 113 K. pneumoniae including 56 ESBL- and 57 non ESBL-producers were collected from six hospitals located in Bushehr province, in the south of Iran, during a period spanning from November 2017 to February 2019. Bacterial isolates were identified using biochemical tests and confirmed by PCR targeting malate dehydrogenase (mdh) the genus-specific housekeeping gene (Fig. 1 and Table 1) (13–15). ESBL producing K. pneumoniae isolates had already been detected by combined disk test (CDT) in our previous study (13).
Fig. 1.
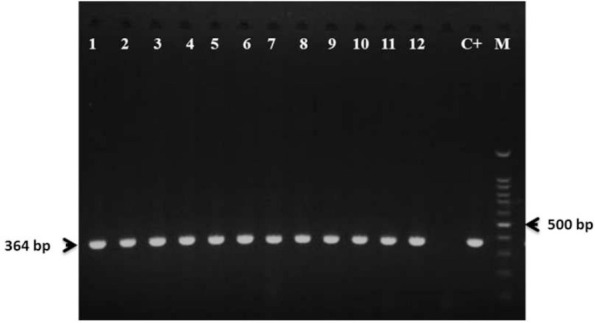
Agarose gel electrophoresis of mdh gene (364 bp) of K. pneumoniae, lanes 1–12: K. pneumoniae clinical isolates carrying mdh gene, C+: K. pneumoniae ATCC 700603, M: 100 bp DNA ladder.
Table 1.
Oligonucleotide primers used for virulence gene detection
| Target gene | Primer Sequence (5′-3′) | Annealing temp. (°C) | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| mdh | F: GCGTGGCGGTAGATCTAAGTCATA | 55 | 364 | (13) |
| R: TTCAGCTCCGCCACAAAGGTA | ||||
| K1(magA) | F: GGTGCTCTTTACATCATTGC | 54 | 1283 | (6, 10) |
| R:GCAATGGCCATTTGCGTTAG | ||||
| K2 | F:GACCCGATATTCATACTTGACAGAG | 60 | 641 | (10, 22) |
| R:CCTGAAGTAAAATCGTAAATAGATGGC | ||||
| rmpA | F:ACTGGGCTACCTCTGCTTCA | 54 | 535 | (23) |
| R:CTTGCATGAGCCATCTTTCA | ||||
| allS | F:CCGTTAGGCAATCCAGAC | 52 | 1091 | (23) |
| R: TCTGATTTAACCCACATT | ||||
| kfu | F:GAAGTGACGCTGTTTCTGGC | 58 | 797 | (23) |
| R:TTTCGTGTGGCCAGTGACTC | ||||
| mrkD | F:GGCGTTGGCGCTCAGATAGG | 57 | 340 | (10) |
| R:AAGCTATCGCTGTACTTCCGGCA | ||||
| fimH | F:GCGCTGGCCGATACCACCACGG | 60 | 423 | (23) |
| R:GCGTAGTAACGCGCCTGGAACGG | ||||
| aero | F:GCATAGGCGGATACGAACAT | 55 | 556 | (10) |
| R:CACAGGGCAATTGCTTACCT | ||||
| ybtS | F:GACGGAAACAGCACGGTAAA | 54 | 242 | (10) |
| R:GAGCATAATAAGGCGAAAGA | ||||
| entB | F:GTCAACTGGGCCTTTGAGCCGTC | 62 | 400 | (10) |
| R:TATGGGCGTAAACGCCGGTGAT | ||||
| iutA | F:GGGAAAGGCTTCTCTGCCAT | 57 | 920 | (10) |
| R:TTATTCGCCACCACGCTCTT | ||||
| wcaG | F:GGTTGGTTCAGCAATCGTA | 53 | 168 | (10) |
| R:ACTATTCCGCCAACTTTTGC | ||||
| wabG | F:ACCATCGGCCATTTGATAGA | 55 | 683 | (23) |
| R:CGGACTGGCAGATCCATATC |
Phenotypic characterization of virulence factors. All ESBL and non-ESBL producing K. pneumoniae clinical isolates were examined for their presence of various virulence factors phenotypically. The following virulence factors like hypermucoviscosity, haemolysin production, protease activity, gelatinase production, lipase production, lecithinase activity, and biofilm formation were tested for all K. pneumoniae isolates by standard methods as follows.
String test for hypermucoviscosity. The hyperviscosity phenotype of the K. pneumoniae isolates was determined using a modified string test. Briefly, a bacteriologic loop was used to stretch a mucoviscous string from a colony of K. pneumoniae grown over night on a blood agar plate at 37°C. Formation of viscous string > 5 mm in length was considered as presence of hypermucoviscosity among the tested isolates (16).
Hemolysin production. For the hemolytic activity assay, an overnight K. pneumoniae culture was streaked on 5% Sheep blood agar following overnight incubation at 37°C. The presence of hemolysis surrounding the colonies was documented (17).
Protease activity. Protease activity assay was carried out to detect K. pneumoniae proteases which are able to degrade casein. From each K. pneumoniae clinical isolate a single colony was picked up from the primary TSA medium and ‘spot’ inoculated on Nutrient agar containing 3% skimmed milk at 37°C for 72 hours. Protease production was identified by the formation of a clear zone around the colonies (12, 17).
Gelatinase activity. To detect K. pneumoniae gelatinase that is able to degrade gelatin, a single colony of fresh bacterial culture was spot-inoculated on Nutrient agar plates containing 3% gelatin. Inoculated plates were incubated at 37°C for 16 hours followed by 4°C for 5 hours. Gelatinase producing isolates were identified by demonstrating a zone of turbid halo around the colonies (17, 18).
Lecithinase activity. This assay was carried out to check for the production of lecithinase (phospholipase C). K. pneumoniae isolates grown overnight on TSA were inoculated onto Egg yolk agar at 37°C up to 7 days. A white precipitate around or beneath an inoculum spot indicated lecithinase formation (12).
Lipase activity. Trypticase soy agar plates supplemented with 1% Tween 80 were inoculated with each tested isolate. After a week of incubation at 37°C, lipase producing isolates form an opaque precipitation zones surrounding or under the inocula (12, 19).
Quantitative biofilm formation assay. The microtiter-plate test is the most frequently used technique for quantifying biofilm formation in which the bacterial film lining the microtiter-plate test is stained with a cationic dye and the optical density (OD) of the stained bacterial film is determined spectrophotometrically (20). Briefly, for each strain, three wells of a 96-well flat-bottomed plastic tissue culture plate (Wuxi Nest Biotechnology, China) were filled with 180 μl of LB supplemented with 1% glucose and 20 μl of the overnight culture adjusted to 0.5 McFarland. As a negative control, sterile LB supplemented with 1% glucose without bacteria was used. The plate was covered with a lid and incubated for 18 h at 37°C. Then, the content of each well was removed, and the wells were carefully washed three times with 200 μL of sterile PBS. The plate was dried for 1 hour at 60°C and stained for 15 min with 180 μl of 0.1% Hucker’s crystal violet (Merck, Germany). The excess stain was rinsed off by rinsing the plate under tap water, and the plate was dried for 10 min at 60°C. The dye bound to the adherent cells was solubilized with 180 μl of 33% (v/v) glacial acetic acid per well. The optical density (OD) of each well was measured at 570 nm using a microplate photometer (Multi scan FC; Thermo Scientific). Each assay was performed in triplicate and repeated 3 times. The OD cut-off (ODc) was defined as three standard deviations above the mean OD of the negative control. All the strains were classified on the basis of the adherence ability into the following categories: non-biofilm producers (OD ≤ ODc), weak biofilm producers (ODc < OD ≤ 2×ODc), moderate biofilm producers (2×ODc < OD ≤ 4×ODc), and strong biofilm producers (4×ODc < OD) (2, 20, 21).
Detection of virulence-associated genes. Multiple genes have been presumed associated with increased virulence in K. pneumoniae strains, hence polymerase chain reaction analysis was used to detect the following virulence genes: fimberial adhesins (fimH-1 (type 1 fimbriae) and mrkD (type 3 fimbriae)), iron-acquisition systems (entB (enterobactin biosynthesis), aero (aerobactin), ybtS (yersiniabactin biosynthesis) and kfu (responsible for an iron uptake system)), gene codifying for capsule (K1 or magA (mucoviscosity-associated gene A), K2, wcaG), rmpA (regulator of mucoid phenotype A)), allS (associated with allantoin metabolism), and wabG (involved in the biosynthesis of the outer core lipopolysaccharide).
Total DNA was extracted using an extraction kit (GeneAll, Korea) as recommended by the manufacturer. The 25 μl reaction mixture is composed of 12.5 μl PCR MasterMix (2x) (Ampliqon, Odense, Denmark), 1 μl of forward primer (10 μM), 1 μl of reverse primer (10 μM), 1 μl of bacterial DNA (50–100 ng), and 9.5 μl of nuclease-free water. The primers and PCR conditions used to assess the prevalence of virulence genes in K. pneumoniae isolates are listed in Table 1. The PCR program consisted of an initial denaturation step at 94°C for 5 min, 30 cycles of DNA denaturation at 94°C for 30 s, primer annealing at a specific annealing temperature (Table 1) for 30 s, and primer extension at 72°C for 40 s followed by a final extension step at 72°C for 5 min. The amplified products were visualized by gel documentation system (Upland, CA, USA) after electrophoresis on 1% agarose gel stained with safe dye (Yekta Tajhiz Azma, Iran). To confirm the PCR results, amplicons were purified and sequenced by the Bioneer Company (Seoul, Korea). Sequences were analyzed using BLAST online through the National Center for Biotechnology Information database.
Statistical analysis. The χ2 and Fisher’s exact tests were used wherever appropriate for statistical evaluation to compare the different variables between ESBL and non-ESBL producing K. pneumoniae isolates. A P value of ≤ 0.05 was considered to be statistically significant. The statistical software used was SPSS for MS Windows, version 24.0.1 (SPSS, Chicago, IL).
Nucleotide sequence accession number. The sequences of virulence genes were submitted to the GenBank database under accession numbers ON087544-ON087555 and ON067807.
RESULTS
Bacterial isolation and identification. One hundred and thirteen clinical isolates of K. pneumoniae were collected from six hospitals located in Bushehr province, in the south of Iran, during a period spanning from November 2017 to February 2019.
Among 113 isolates 56 were ESBL producers and 57 were non ESBL-producers (13). The majority of isolates were cultured from urine (n=86, 76.1%) followed by endotracheal tube (ETT) secretions (n=11, 9.7%), wound (n=5, 4.4%), blood (n=4, 3.5%), burn (n=3, 2.7%), ascites (n=1, 0.9%), liver abscess (n=1, 0.9%), sputum (n=1, 0.9%), and shaldon catheter (n=1, 0.9%).
Phenotypic characterization of virulence factors of K. pneumoniae isolates. Among various phenotypic virulence factors tested, hemolysin production, biofilm formation, protease activities, hypermocoviscosity and lipase production in 113 K. pneumoniae clinical isolates were observed in 100%, 76.9%, 62.9%, 13.3% and 2.7% of isolates, respectively (Fig. 2). None of the tested isolates showed neither gelatinase nor lecithinase activity.
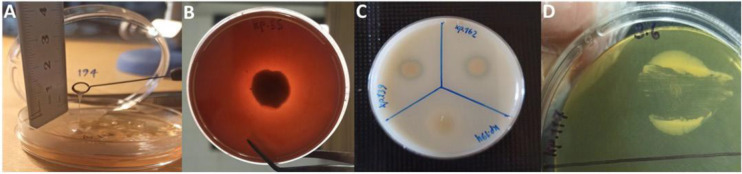
Fig. 2.
Phenotypic detection of virulence factors of K. pneumoniae clinical isolates. A: Formation of viscous string > 5 mm in length as a result of positive string test, B: Presence of haemolysis surrounding the colonies by hemolysin production, C: formation of clear zone around the colonies as a result of protease activity and D: Opaque precipitation under the inocula by lipase activity.
Protease activity was observed in 34% of ESBL-producing isolates and 37% of non-ESBL-producers. For lipase production only 2 (3.6%) ESBL and 1 (1.8%) non-ESBL producing isolates were considered lipase producers. There was no statistically significant difference of both groups in protease and lipase production. The prevalence of hypermocoviscosity phenotype was lower among ESBLs producing isolates where 7.1% (n=4) of ESBLs exhibited hypermucoviscosity compared to non-ESBLs (19.3%, n= 11) (P =0.057). The phenotypic virulence features of 56 ESBLs and 57 non-ESBLs producing isolates are shown in Tables 2 and 3. The number of hypermucoviscous isolates was higher among non-ESBL producers.
Table 2.
Comparison of the phenotypic virulence factors between 56 ESBL and 57 non ESBL K. pneumoniae isolates.
| Virulence factor | ESBL, No. (%) | Non-ESBL, No. (%) | p-value |
|---|---|---|---|
| Hypermocoviscosity | 4 (7.1) | 11 (19.3) | 0.057 |
| Hemolysin | 56 (100) | 57 (100) | _ |
| Protease | 34 (60.7) | 37 (64.9) | 0.64 |
| Lipase | 2 (3.6) | 1 (1.8) | 0.61 |
| Gelatinase | 0 | 0 | _ |
| Lecithinase | 0 | 0 | _ |
Table 3.
Comparison of biofilm intensity between 56 ESBL and 57 Non ESBL K. pneumoniae isolates.
| ESBL, No. (%) | Non ESBL, No. (%) | Total | ||
|---|---|---|---|---|
| Biofilm | Non | 12 (21.4) | 14 (24.6) | 26 (23) |
| Weak | 16 (28.6) | 34 (59.6) | 50 (44.2) | |
| Moderate | 24 (42.9) | 9 (15.8) | 33 (29.2) | |
| Strong | 4 (7.1) | 0 (0) | 4 (3.5) | |
| Total | 56 (100) | 57 (100) | 113 (100) |
The ability to produce biofilm is another important virulence factor in K. pneumoniae. The microtiter plate assay was used to measure biofilm production in all isolates. Biofilm intensity was categorized as weak, moderate and strong and comparisons were made among ESBL and non-ESBL producers. In this study, 77% (n = 87) of all K. pneumoniae isolates including 78.6% (n=44) of ESBL and 75.4 % (n=43) of non-ES-BL producers demonstrated biofilm-forming ability in the in vitro assay and there was no statistically significant difference in biofilm formation between ESBL and non-ESBL producers (P = 0.69).
Weak biofilm was observed in 59.6% of non-ESBL producers and 28.6% of ESBL producers. Moderate biofilm turned out to be higher in ESBL (42.9%) in comparison to non-ESBLs (15.8%) and strong biofilm production was only detected in 7.1% of ESBL producers. In addition, 12 (21.4%) ESBL producers and 14 (24.6%) non-ESBL producers were non-biofilm producers. It is noteworthy that the intensity of biofilm production (the total number of moderate and strong biofilm producers) was higher in the ESBL producer (28 isolates) compared to the non-ESBL producer (9 isolates) (Table 3).
Detection of virulence genes of K. pneumoniae isolates. The virulence genes including rmpA (4.42%), aero (5.30%), kfu (33.62%), allS (3.53%), mrkD (100%), ybtS (38.93%), entB (99.11%), iutA (5.30%), fimH (100), wabG (100%), wcaG (5.3%), K1, (1.76%), and K2 (6.19%) were detected in our K. pneumoniae isolates (Fig. 3).
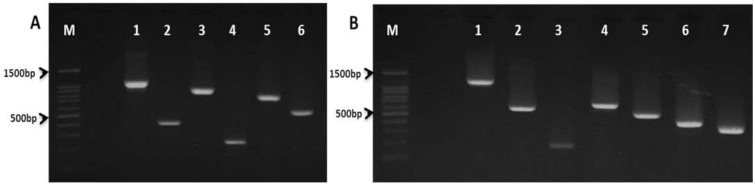
Fig. 3.
Agarose gel electrophoresis of K. pneumoniae virulence genes. A. M: 100 bp DNA ladder.; lane 1: allS gene (1091bp); lane 2: entB (400bp); lane 3: iutA (920bp); lane 4: ybtS (242bp); lane 5: kfu (797bp) and lane 6: aero (556bp). B. M: 100 bp DNA ladder; lane 1: k1 (magA) gene (1283bp); lane 2: k2 (641bp); lane 3: wcaG (168bp); lane 4: wabG (683bp); lane 5: rmpA (535bp); lane 6: fimH (425bp) and lane 7: mrkD (340bp).
Virulence genes mrkD, fimH and wabG were detected in all ESBL and non-ESBL producing isolates while iutA and aero were observed only in 10.5% of non-ESBL-producing isolates and these two genes were not detected in ESBL isolates with significant differences (p≤0.05). Of note, K1, and rmpA gene was also detected only in non-ESBL producer (p=0.057, this borderline P value could be due to the small sample size) (Table 4). There was no significant difference in the presence of other virulence genes between ESBL and non ESBL K. pneumoniae isolates.
Table 4.
Comparison of virulence genes between 56 ESBL and 57 Non ESBL K. pneumoniae isolates.
| Virulence gene | ESBL, No. (%) | NON-ESBL, No. (%) | p-value |
|---|---|---|---|
| mrkD | 56 (100) | 57 (100) | _ |
| fimH | 56 (100) | 57 (100) | _ |
| allS | 1 (1.8) | 4 (7) | 0.36 |
| entB | 56 (100) | 56 (98.2) | 1.000 |
| K1 | 0 (0) | 2 (3.5) | 0.49 |
| rmpA | 0 (0) | 5 (8.8) | 0.057 |
| K2 | 4 (7.1) | 3 (5.3) | 0.71 |
| kfu | 19 (33.3) | 19 (33.9) | 0.94 |
| wcaG | 1 (1.8) | 5 (8.8) | 0.20 |
| iutA | 0 (0) | 6 (10.5) | 0.027 |
| aero | 0 (0) | 6 (10.5) | 0.027 |
| ybtS | 20 (35.7) | 24 (42.1) | 0.48 |
| wabG | 56 (100) | 57 (100) | _ |
In addition, the carriage of allS, K1(magA), K2, rmpA, iutA and aero virulence genes was higher in hypermucoviscous compared to non hypermucoviscous isolates with significance of differences (p≤ 0.05) (Table 5).
Table 5.
Comparison of virulence genes between hypermucoviscous (HM) and non hypermucoviscous (Non-HM) K. pneumoniae isolates.
| Virulence gene | HM, No. (%) | Non-HM, No. (%) | p-value |
|---|---|---|---|
| mrkD | 15 (100) | 98 (100) | _ |
| fimH | 15 (100) | 98 (100) | _ |
| allS | 3 (20) | 2 (2) | 0.016 |
| entB | 14 (93.3) | 98 (100) | 0.13 |
| K1 | 2 (13.3) | 0 (0) | 0.017 |
| rmpA | 5 (33.3) | 0 (0) | 0.0001 |
| K2 | 5 (33.3) | 2 (2) | 0.0001 |
| kfu | 7 (46.7) | 31 (31.6) | 0.25 |
| wcaG | 2 (13.3) | 4 (4.1) | 0.18 |
| iutA | 6 (40) | 0 (0) | 0.0001 |
| aero | 6 (40) | 0 (0) | 0.0001 |
| ybtS | 4 (26.7) | 40 (40.8) | 0.29 |
| wabG | 15 (100) | 98 (100) | _ |
DISCUSSION
Klebsiella pneumoniae is a Gram-negative opportunistic pathogen capable of causing a spectrum nosocomial and community-acquired infections (14). Its pathogenicity depends on a variety of virulence factors, including capsule, fimbriae, iron acquisition systems as well as biofilm formation, and the increasing acquisition of antibiotic resistance such as ESBLs encoding plasmids (24). To determine the virulence potential of a pathogen, it is necessary to detect its virulence factors.
In this study, the presence of various virulence factors including haemolysin production, protease activity, gelatinase production, lipase production, lecithinase activity and hypermucoviscosity was investigated via phenotypic methods. One of the virulence factors among Gram-negative bacteria is the production of hemolysin. Although Klebsiella is known as a non-hemolytic bacterium (25) as stated in studies conducted by Pereira et al. (26) and Barati et al., none of their K. pneumoniae isolates showed hemolytic activity (14). Hemolysin production was observed in 100% of clinical isolates in our study which is higher than the results reported by Gundogan et al. (67%) (25).
Protease plays a significant role in host immune evasion, invasiveness, and tissue damage (12). Of note, 62.8% of our isolates exhibited protease activity. Lipase activity, however, was found only in 2.6% of isolates. Similar to our study, Gharrah et al., have confirmed that 8% of Klebsiella isolates exhibited lipase activity. In this study, none of the isolates were able to produce either lecithinase or gelatinase. It is notable that among the various phenotypic virulence factors investigated in this study, there was no significant difference between ESBL and non-ESBL producers with the exception of hypermucoviscosity phenotype.
Previous studies have reported that a negative association exists between hypermucoviscosity (HV) and ESBL. Indeed, the hypermucoviscosity phenotype was more frequently detected in non-ESBL producers. Furthermore, ESBL genes are hardly ever found in hypermucoviscous K. pneumoniae isolates (27).
In a previous study by Yu et al. 8.8% of ESBL and 53.8% of non-ESBL producers were hypermucoviscous (28). Our results of the hypermucoviscosity revealed that among 113 isolates, 15 isolates (13.3%) showed hypermucoviscous phenotype, of which 5, 2, and 8 isolates belonged to K2, K1, and non K1/K2 capsular serotypes, respectively. The prevalence of hypermucoviscosity in our isolates was consistent with the studies conducted by Rastegar et al. in Kerman, Iran (29) and Taraghian et al. in Isfahan, Iran (30).
In agreement with previous reports, the prevalence of the hypermocoviscosity phenotype was lower among our ESBLs producing isolates where 7.1% of ESBL producers showed hypermucoviscosity in comparison to non-ESBLs (19.3%) (P =0.057, this borderline P value could be due to the small sample size).
Biofilm formation has a great impact on K. pneumoniae pathogenicity. In addition, biofilm producers are less exposed to antimicrobial agents when compared with non-biofilm formers (9). In the current study, 76.9% of K. pneumoniae were biofilm producers which was similar to the other studies conducted in Iran (5, 31). Although biofilm formation was not significantly different between ESBLs and non-ESBLs groups (P = 0.69), the intensity of biofilm (moderate and strong) was clearly higher in ESBLs group as 50% of ESBL producers developed strong and moderate biofilm while only 15.8% of non-ES-BL producers were able to form strong and moderate biofilm. Similar findings were reported by Shadkam et al. (31). Among all tested isolates only 3.5% were strong biofilm producer, while unlike our study, the percentage of strong biofilm producing isolates reported by Shadkam et al. (Sari, Iran) was 25% (31).
Moreover, in the present study virulence genes including plasmid-born rmpA, aerobactin, kfu, allS, mrkD, ybtS, entB, iutA, fimH, wabG, wcaG genes, K1 (magA) and K2 capsular serotype were detected by PCR. All isolates including ESBL and non-ESBL producing isolates harbored fimH, mrkD and wabG genes, and entB gene was detected in 99.1% of isolates. The high percentage of fimH, mrkD and, wabG genes has been reported by other researchers (32–34). It is noteworthy that iutA, aero, rmpA and K1 genes were observed only in non-ESBL-producing isolates. In addition, allS, K1(magA), rmpA, iutA and aero virulence genes were only detected in hypermucoviscous K. pneumoniae isolates and the carriage of K2 gene was higher in hypermucoviscous K. pneumoniae compared to non hypermucoviscous isolates with significance of differences (p≤ 0.05). These findings were similar to the previous studies (7, 35–37).
It has been shown that isolates harboring K1/K2 capsular serotypes are more virulent (38). In the present study, 9 isolates exhibited K1/K2 capsular serotypes, 78% of which were hypermucoviscous. Notably, in this study 81.8% of ETT, 80% of blood, and 75.5% of urine samples were biofilm producers while there was only one sample of liver abscess, which was neither hypermucoviscous nor able to form biofilm but harbored mrkD, fimH, wabG, kfu and entB virulence genes. We found that the most studied virulence genes were identified in 4 hypermucoviscous isolates; Kp194 isolated from urine harbored 12 out of 13 virulence genes, Kp50 harbored 11 virulence genes and 9 virulence genes were detected in Kp19 (ETT) and Kp139 (urine). Of note, Kp194 and kp50 belonged to K1 capsular serotype and Kp19 and Kp139 belonged to K2 capsular serotype.
The high prevalence of ybtS and entB genes of iron acquisition systems in the present study was consistent with studies conducted by Soltani (Tabriz, Iran) (39) and Bachman (40) (Pennsylvania, USA). Unlike the prevalence of ybtS and entB genes, only 6 isolates (5.30%) carried the aero gene. All these 6 isolates were hypermucoviscous. Five out of 6 isolates belong to K1/K2 serotypes and only one isolate belongs to non K1/K2 capsular serotypes. In Yu et al.’s study, 90% of isolates harbored aero gene, which was not consistent with our study (7). This significant discrepancy with our study might also be due to the source of their samples, which were recovered from liver abscesses.
CONCLUSION
Among phenotypic virulence factors, biofilm intensity was more common in ESBL producers while the hypermucoviscosity phenotype was more frequently detected in non-ESBL producing isolates. Genotypic investigation of virulence genes revealed that iutA, aero, rmpA and K1 genes were observed only in non-ESBL-producing isolates. In addition, allS, K1(magA), rmpA, iutA and aero virulence genes were only detected in hypermucoviscous K. pneumoniae isolates and K2 gene was higher in hypermucoviscous compared to non hypermucoviscous isolates. Moreover, beside the biofilm development potential in ESBL-producing isolates, this report confirms the presence of hypervirulent K. pneumoniae strains among isolates of Bushehr province. Therefore, the identification of potentially pathogenic isolates plays an important role in preventing their spread as well as the success of their treatment.
ACKNOWLEDGEMENTS
This article was from the postgraduate MSc thesis of Hamed Hatami Mirbag and was supported by the Vice-Chancellor of Research of Bushehr University of Medical Sciences, Bushehr, Iran (grant no. 1449). We would like to thank Clinical Research Development Center, The Persian Gulf Martyrs Hospital, Bushehr University of Medical Sciences, Bushehr, Iran for facilitating the process of sampling.
REFERENCES
- 1.Kim YJ, Kim SI, Kim YR, Wie SH, Lee HK, Kim S-Y, et al. Virulence factors and clinical patterns of hypermucoviscous Klebsiella pneumoniae isolated from urine. Infect Dis (Lond) 2017; 49: 178–184. [DOI] [PubMed] [Google Scholar]
- 2.Vuotto C, Longo F, Pascolini C, Donelli G, Balice MP, Libori MF, et al. Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains. J Appl Microbiol 2017; 123: 1003–1018. [DOI] [PubMed] [Google Scholar]
- 3.Cubero M, Marti S, Domínguez MÁ, González-Díaz A, Berbel D, Ardanuy C. Hypervirulent Klebsiella pneumoniae serotype K1 clinical isolates form robust biofilms at the air-liquid interface. PLoS One 2019; 14(9): e0222628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu VL, Hansen DS, Ko WC, Sagnimeni A, Klugman KP, Von Gottberg A, et al. Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg Infect Dis 2007; 13: 986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mirzaie A, Ranjbar R. Antibiotic resistance, virulence-associated genes analysis and molecular typing of Klebsiella pneumoniae strains recovered from clinical samples. AMB Express 2021; 11: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu W-L, Fung C-P, Ko W-C, Chuang Y-C, Lee C-C, Chuang Y-C. Polymerase chain reaction analysis for detecting capsule serotypes K1 and K2 of Klebsiella pneumoniae causing abscesses of the liver and other sites. J Infect Dis 2007; 195: 1235–1236. [DOI] [PubMed] [Google Scholar]
- 7.Yu W-L, Ko W-C, Cheng K-C, Lee C-C, Lai C-C, Ch- uang Y-C. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis 2008; 62: 1–6. [DOI] [PubMed] [Google Scholar]
- 8.Wasfi R, Elkhatib WF, Ashour HM. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci Rep 2016; 6: 38929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Surgers L, Boyd A, Girard P-M, Arlet G, Decré D. Biofilm formation by ESBL-producing strains of Escherichia coli and Klebsiella pneumoniae. Int J Med Microbiol 2019; 309: 13–18. [DOI] [PubMed] [Google Scholar]
- 10.Fu L, Huang M, Zhang X, Yang X, Liu Y, Zhang L, et al. Frequency of virulence factors in high biofilm formation blaKPC-2 producing Klebsiella pneumoniae strains from hospitals. Microb Pathog 2018; 116: 168–172. [DOI] [PubMed] [Google Scholar]
- 11.Lee S, Han SW, Kim KW, Song DY, Kwon KT. Third-generation cephalosporin resistance of community-onset Escherichia coli and Klebsiella pneumoniae bacteremia in a secondary hospital. Korean J Intern Med 2014; 29: 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas R, Hamat RA, Neela V. Extracellular enzyme profiling of Stenotrophomonas maltophilia clinical isolates. Virulence 2014; 5: 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latifi B, Tajbakhsh S, Ahadi L, Yousefi F. Coexistence of aminoglycoside resistance genes in CTX-M-producing isolates of Klebsiella pneumoniae in Bushehr province, Iran. Iran J Microbiol 2021; 13: 161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barati A, Ghaderpour A, Chew LL, Bong CW, Thong KL, Chong VC, et al. Isolation and characterization of aquatic-borne Klebsiella pneumoniae from tropical estuaries in Malaysia. Int J Environ Res Public Health 2016; 13: 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thong KL, Lai MY, Teh C SJ, Chua KH. Simultaneous detection of methicillin-resistant Staphylococcus aureus, Acinetobacter baumannii, Escherichia coli, Klebsiella pneumoniae and Pseudomonas aeruginosa by multiplex PCR. Trop Biomed 2011; 28: 21–31. [PubMed] [Google Scholar]
- 16.Wang T-C, Lin J-C, Chang J-C, Hiaso Y-W, Wang C-H, Chiu SK, et al. Virulence among different types of hypervirulent Klebsiella pneumoniae with multi-locus sequence type (MLST)-11, Serotype K1 or K2 strains. Gut Pathog 2021; 13: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalaivani R, Jenitha JP, Seetha KS. Clinical isolates of Klebsiella pneumoniae and its virulence factors from a tertiary care hospital. Indian J Microbiol Res 2019; 6: 109–112. [Google Scholar]
- 18.Cevahir N, Demir M, Kaleli I, Gurbuz M, Tikvesli S. Evaluation of biofilm production, gelatinase activity, and mannose-resistant hemagglutination in Acinetobacter baumannii strains. J Microbiol Immunol Infect 2008; 41: 513–518. [PubMed] [Google Scholar]
- 19.Khalil MA, Ibrahim Sonbol F, Mohamed AF, Ali SS. Comparative study of virulence factors among ES-βL-producing and nonproducing Pseudomonas aeruginosa clinical isolates. Turk J Med Sci 2015; 45: 60–69. [DOI] [PubMed] [Google Scholar]
- 20.Stepanović S, Vuković D, Dakić I, Savić B, Švabić-Vlahović M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods 2000; 40: 175–179. [DOI] [PubMed] [Google Scholar]
- 21.Donelli G, Vuotto C, Cardines R, Mastrantonio P. Biofilm-growing intestinal anaerobic bacteria. FEMS Immunol Med Microbiol 2012; 65: 318–325. [DOI] [PubMed] [Google Scholar]
- 22.Lan Y, Zhou M, Jian Z, Yan Q, Wang S, Liu W. Prevalence of pks gene cluster and characteristics of Klebsiella pneumoniae-induced bloodstream infections. J Clin Lab Anal 2019; 33(4): e22838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiang T-T, Yang Y-S, Yeh K-M, Chiu S-K, Wang N-C, Lin T-Y, et al. Quantification and comparison of virulence and characteristics of different variants of carbapenemase-producing Klebsiella pneumoniae clinical isolates from Taiwan and the United States. J Microbiol Immunol Infect 2016; 49: 83–90. [DOI] [PubMed] [Google Scholar]
- 24.Swathi CH, Chikala R, Ratnakar KS, Sritharan V. A structural, epidemiological & genetic overview of Klebsiella pneumoniae carbapenemases (KPCs). Indian J Med Res 2016; 144: 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gundogan N, Citak S, Yalcin E. Virulence properties of extended spectrum β-lactamase-producing Klebsiella species in meat samples. J Food Prot 2011; 74: 559–564. [DOI] [PubMed] [Google Scholar]
- 26.Pereira SC, Vanetti MC. Potential virulence of Klebsiella sp. isolates from enteral diets. Braz J Med Biol Res 2015; 48: 782–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hennequin C, Robin F. Correlation between antimicrobial resistance and virulence in Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis 2016; 35: 333–341. [DOI] [PubMed] [Google Scholar]
- 28.Yu W-L, Lee M-F, Tang H-J, Chang M-C, Chuang Y-C. Low prevalence of rmpA and high tendency of rmpA mutation correspond to low virulence of extended spectrum β-lactamase-producing Klebsiella pneumoniae isolates. Virulence 2015; 6: 162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rastegar S, Moradi M, Kalantar-Neyestanaki D, Ali Golabi D, Hosseini-Nave H. Virulence factors, capsular serotypes and antimicrobial resistance of hypervirulent Klebsiella pneumoniae and classical Klebsiella pneumoniae in Southeast Iran. Infect Chemother 2019; 10.3947/ic.2019.0027. [DOI] [PubMed] [Google Scholar]
- 30.Taraghian A, Nasr Esfahani B, Moghim S, Fazeli H. Characterization of hypervirulent extended-spectrum β-lactamase-producing Klebsiella pneumoniae among urinary tract infections: the first report from Iran. Infect Drug Resist 2020; 13: 3103–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shadkam S, Goli HR, Mirzaei B, Gholami M, Ahanjan M. Correlation between antimicrobial resistance and biofilm formation capability among Klebsiella pneumoniae strains isolated from hospitalized patients in Iran. Ann Clin Microbiol Antimicrob 2021; 20: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El Fertas-Aissani R, Messai Y, Alouache S, Bakour R. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol Biol (Paris) 2013; 61: 209–216. [DOI] [PubMed] [Google Scholar]
- 33.Hasani A, Soltani E, Ahangarzadeh Rezaee M, Pirzadeh T, Ahangar Oskouee M, Hasani A, et al. Serotyping of Klebsiella pneumoniae and its relation with capsule-associated virulence genes, antimicrobial resistance pattern, and clinical infections: a descriptive study in medical practice. Infect Drug Resist 2020; 13: 1971–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bakhtiari R, Javadi A, Aminzadeh M, Molaee-Aghaee E, Shaffaghat Z. Association between presence of rmpA, mrkA and mrkD Genes and antibiotic resistance in clinical Klebsiella pneumoniae isolates from hospitals in Tehran, Iran. Iran J Public Health 2021; 50: 1009–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dan B, Dai H, Zhou D, Tong H, Zhu M. Relationship between drug resistance characteristics and biofilm formation in Klebsiella Pneumoniae strains. Infect Drug Resist 2023; 16: 985–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung SW, Chae HJ, Park YJ, Yu JK, Kim SY, Lee HK, et al. Microbiological and clinical characteristics of bacteraemia caused by the hypermucoviscosity phenotype of Klebsiella pneumoniae in Korea. Epidemiol Infect 2013; 141: 334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu W-L, Ko W-C, Cheng K-C, Lee H-C, Ke D-S, Lee C-C, et al. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis 2006; 42: 1351–1358. [DOI] [PubMed] [Google Scholar]
- 38.Fu Y, Xu M, Liu Y, Li A, Zhou J. Virulence and genomic features of a blaCTX-M-3 and blaCTX-M-14 coharboring hypermucoviscous Klebsiella pneumoniae of serotype K2 and ST65. Infect Drug Resist 2019; 12: 145–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soltani E, Hasani A, Rezaee MA, Pirzadeh T, Oskouee MA, Hasani A, et al. Virulence characterization of Klebsiella pneumoniae and its relation with ESBL and AmpC beta-lactamase associated resistance. Iran J Microbiol 2020; 12: 98–106. [PMC free article] [PubMed] [Google Scholar]
- 40.Bachman MA, Oyler JE, Burns SH, Caza M, Lépine F, Dozois CM, et al. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect Immun 2011; 79: 3309–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]