Figure 4. Flavin reductases are induced by their electron acceptors and exhibit relatively narrow substrate specificities.
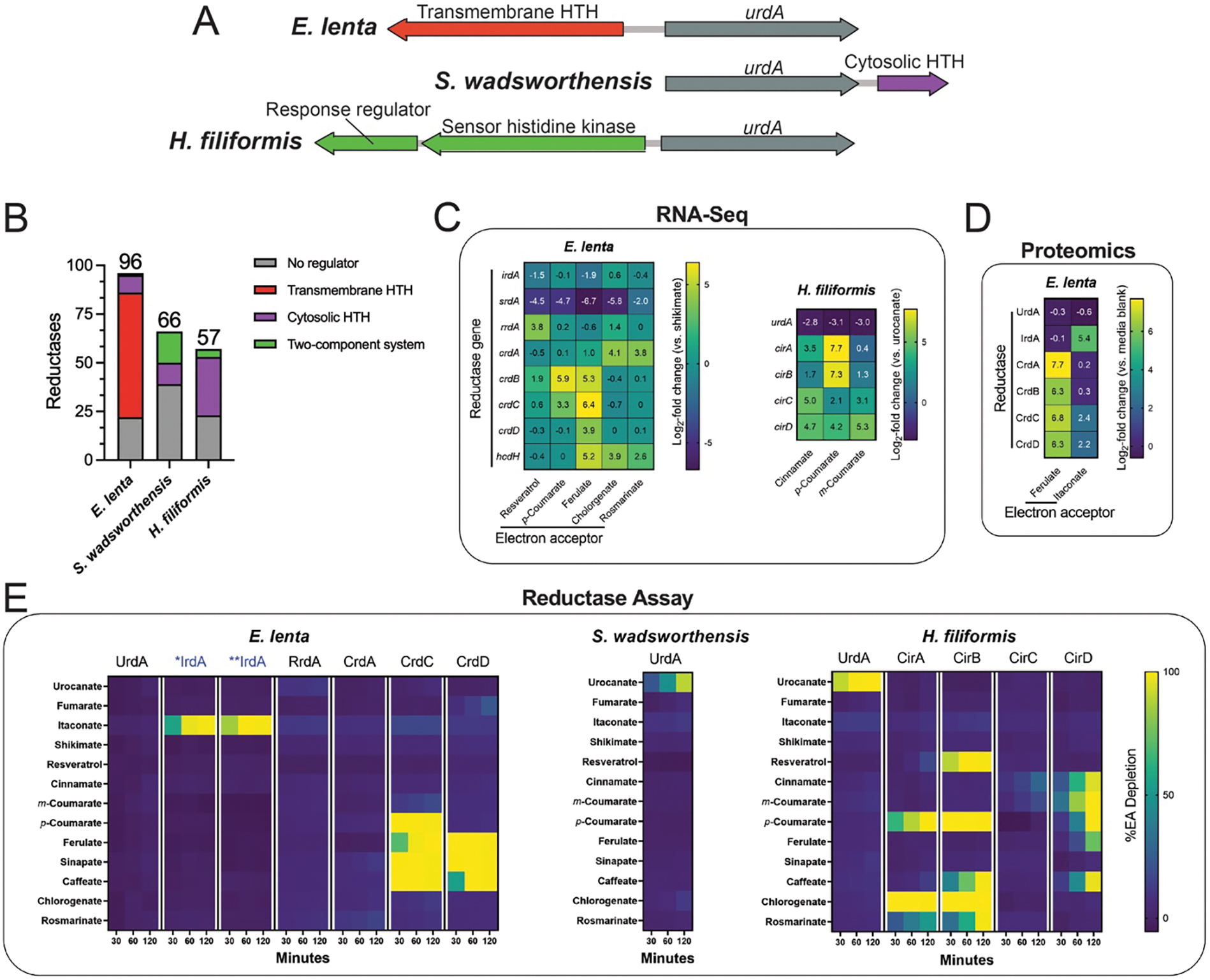
(A) Genomic context of urdA urocanate reductases in E. lenta DSM2243, S. wadsworthensis DFI.4.78, and H. filiformis DFI.9.20 genomes. Genes encoding predicted transcriptional regulators are noted. (B) Genomic context of flavin and molybdopterin reductases with respect to adjacent regulatory elements. Either no clear element present (grey), two-component system (green), cytosolic regulator (purple), or transmembrane regulator (red). Cytosolic and transmembrane refers to the cellular localization of the putative signal-receiving domain in predicted transcriptional regulators that contain a DNA-binding helix-turn-helix (HTH) domain. The number of flavin and molybdopterin reductases genes that directly neighbor a predicted transcriptional regulator in E. lenta DSM2243, S. wadsworthensis DFI.4.78, and H. filiformis DFI.9.20 genomes. (C) RNA-Seq results from E. lenta DSM2243 and H. filiformis DFI.9.20 cells cultivated in media supplemented with indicated electron acceptors. (D) Proteomics results from E. lenta DSM2243 cells cultivated in media supplemented with indicated electron acceptors. (E) Activity of recombinant reductases in the presence of indicated electron acceptors. For (E), representative data are shown.
