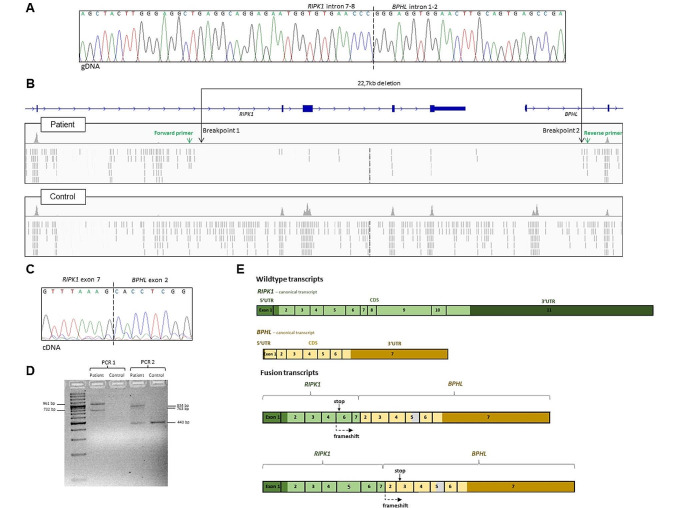
Fig. 3.
Chromatogram and Integrative Genome Viewer (IGV) visualization of the alignment. (A) Amplification of the genomic area carrying the deletion revealed the precise breakpoints of the deletion. Primers: 5’TGAGTTGGAGATTGGGGTGC3’ (forward) and 5’TTATGGGTGCCGTACAGGTG3’ (reverse). (B) The IGV coverage tracks of the patient and control exomes illustrate the position of the homozygous 22,726 bp deletion encompassing exons 8–11 and exon 1 in RIPK1 (NM_001354930.2) and BPHL (NM_004332.4), respectively. The deletion was called from whole-exome data based on coverage values using the ExomeDepth software [10]. Since the breakpoints were located outside the targeted regions, their exact position could not be inferred from the whole-exome data alone. As illustrated by the IGV screenshot, the density of off-target reads mapped to the introns 7–8 (RIPK1) and 1–2 (BPHL) were visibly sparser in the patient compared to controls. Primers were designed based on the assumption that the breakpoint was located near the points where the read density starts to decrease (primer binding sites are indicated with green arrows). (C) Chromatogram showing the breakpoint of the cDNA fusion transcript between RIPK1 and the following gene BPHL. cDNA was synthesized from mRNA from B-cell lines derived from the patient and a healthy control. PCR and Sanger sequencing showed that the deletion leads to a fusion transcript between RIPK1 and BPHL in the patient. (D) Gel electrophoresis of the products from PCR 1 (primers binding in RIPK1 exon 1 and BPHL exon 3; forward:5‘GGAAGGTGTCTCTGTGTTTCCA3’, reverse: 5‘GAGGTCCAAAATCAGTCTCTCCA3‘) and PCR 2 (primers binding in RIPK1 exon 6 and BPHL exon 7; forward: 5‘GCTCTGCTGGGAAGCGAAT3‘, reverse: 5‘GGTTGTGTTTGCCTTCTGGC3‘). Sanger sequencing showed that the multiple bands correspond to different splice variants, caused by skipping of RIPK1 exon 5 (PCR 1, 732 bp product) and the partial skipping of BPHL exon 5 (PCR 2, 763 bp product). The 443 bp band in PCR 2 is due to unspecific amplification of ABCB8 cDNA in both the patient and the control subject. In the healthy donor, no other products were amplified due to the absence of a fusion transcript. (E) Illustration of the wildtype transcripts and the mutant fusion transcripts in the patient. All transcripts identified in the patient are predicted to lead to a frameshift and premature stop in either RIPK1 exon 6 or BPHL exon 3, depending on whether or not RIPK1 exon 5 is skipped. In a proportion of the transcripts, part of BPHL exon 5 is skipped (indicated in grey). Since RIPK1 exon 5 and BPHL exon 5 were not included in the same amplicons, we do not know if the partial BPHL exon 5 skipping occurred in the transcripts including RIPK1 exon 5, excluding RIPK1 exon 5, or both