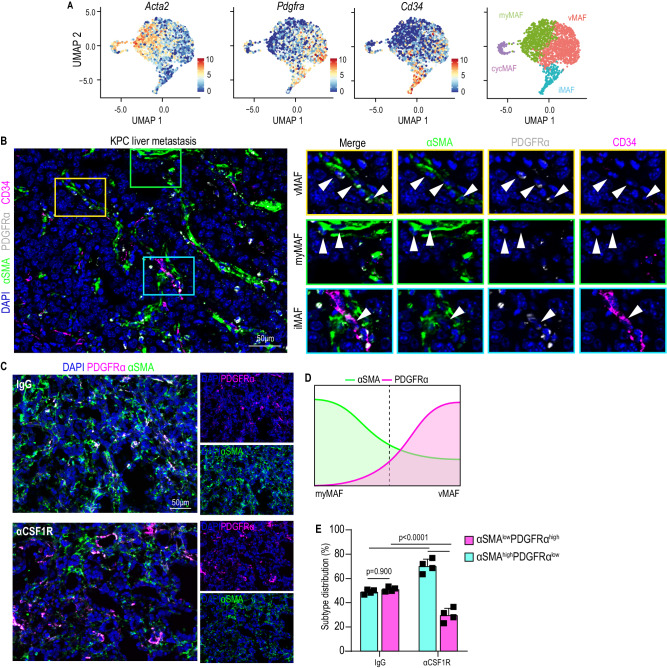
Fig. 2. Transcriptional and spatially diverse MAF populations co-exist in PDAC liver metastasis.
A UMAP plot depicting expression of Acta2, Pdgfra, and Cd34 in vMAF, myMAF, iMAF, and cycMAF clusters. B Representative immunofluorescence image of annotated MAF subtypes in liver metastasis of KPC mice. Individual MAF subtypes are defined on the representative image in coloured regions: vMAF (αSMALow, PDGFRαHigh, CD34Low) – red; myMAF (αSMAHigh, PDGFRαLow, CD34Low) – green; iMAF (αSMALow, PDGFRαHigh, CD34High) – blue). Arrowheads indicate the defined MAF subtype. N = 3 mice. Scale bar, 50 µm. C Representative immunofluorescence image for the distribution of PDGFRα and and αSMA to discern vMAFs and myMAFs in metastatic tumours derived from Pdgfrb-GFP mice treated with IgG or αCSF1R neutralising antibody. To the right, separated channel images are shown. Scale bar, 50 µm. D Representative schematic depicting how the gradated expression of αSMA and PDGFRα separates across vMAF and myMAF subpopulations, as observed in A, is utilised as a strategy to analyse changes in abundance between both subpopulations. E Quantification of the distribution of αSMAlowPDGFRαhigh and αSMAhighPDGFRαlow cells in C. Data is presented as mean percentage distribution averaged from n = 4 mice per group. Error bars, SD. P values, two-way ANOVA with Tukey’s multiple comparisons test. Source data are provided as a Source Data file.