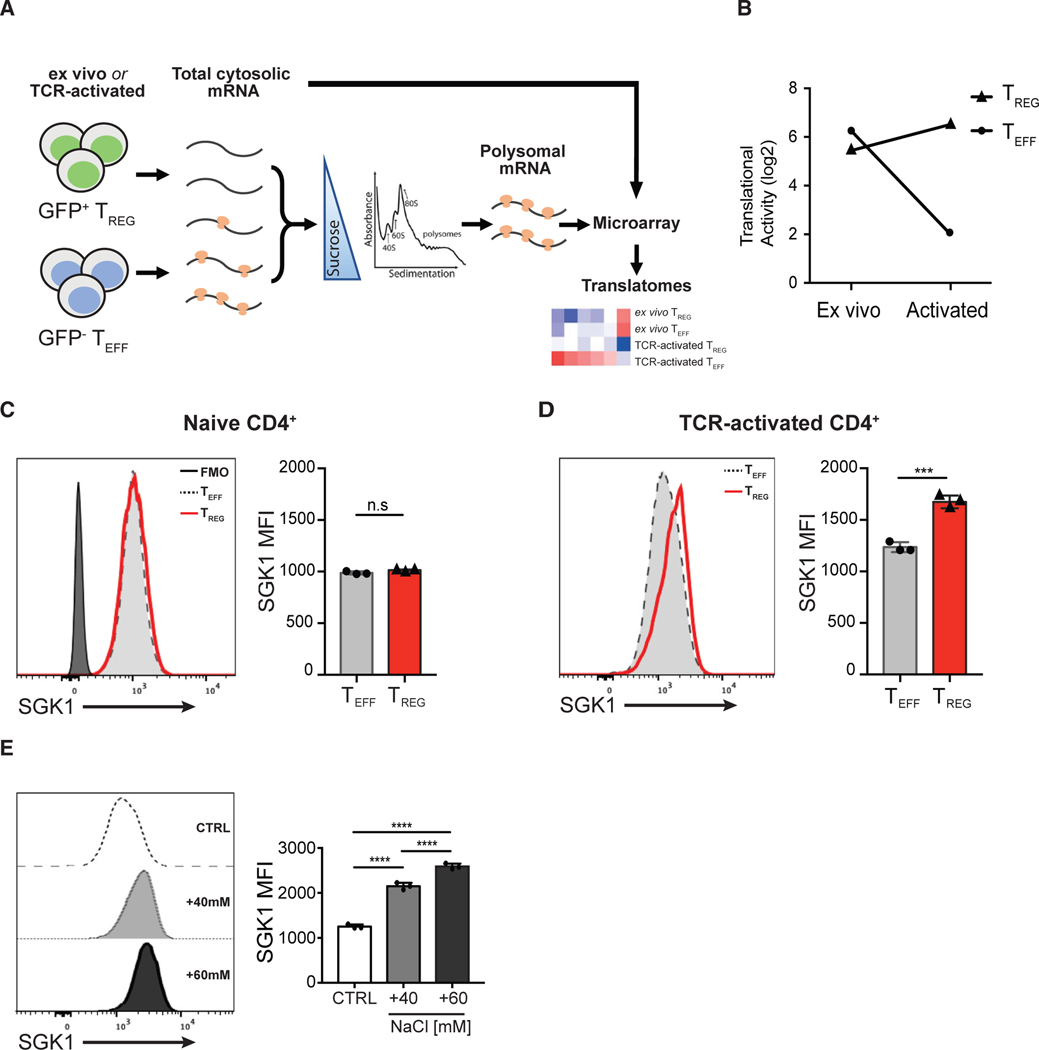
Figure 1. Salt-Sensing Kinase SGK1 mRNA Is Preferentially Translated in Foxp3+ Treg Cells.
(A) Schematic diagram showing the workflow of translatome profiling in CD4+ T cell subsets. FACS-purified Teff and Treg cells that were naive (directly ex vivo) or TCR activated (in vitro) were treated with cycloheximide to immobilize ribosomes on the mRNA. Polysome-associated (≥3 ribosomes) mRNAs were fractionated from total cytosolic mRNA by sedimentation through a sucrose gradient. Total cytosolic mRNA and polysome-associated mRNA were probed with microarrays to quantify mRNA level. Polysome-associated mRNA pools describe the translatomes of CD4+ T cell subsets.
(B) Quantitative representation of SGK1 translational activity (after correction for cytosolic mRNA levels) in CD4+ Teff and Treg cells ex vivo or after in vitro activation. Data are shown as log2 fold change.
(C) Representative flow cytometric histograms and quantitative analysis of SGK1 protein expression in ex vivo CD4+ T cell subsets by intracellular staining of SGK1 and the assessment of geometric mean fluorescent intensity (gMFI).
(D) Representative flow cytometric histograms and quantitative analysis of SGK1 protein expression in in vitro TCR-activated CD4+ T cell subsets by intracellular staining of SGK1 and the assessment of geometric mean fluorescent intensity (gMFI).
(E) Representative flow cytometric histograms and quantitative analysis of SGK1 protein expression in CD4+ T cells treated with increasing concentration of salt through intracellular staining of SGK1 and the assessment of geometric mean fluorescent intensity (gMFI).
Polysomal profiling experiments (A and B) were performed in biological duplicates. Data shown in (D) and (E) are representative of three independent experiments with triplicates of each condition. Error bars represent mean ± standard deviation.