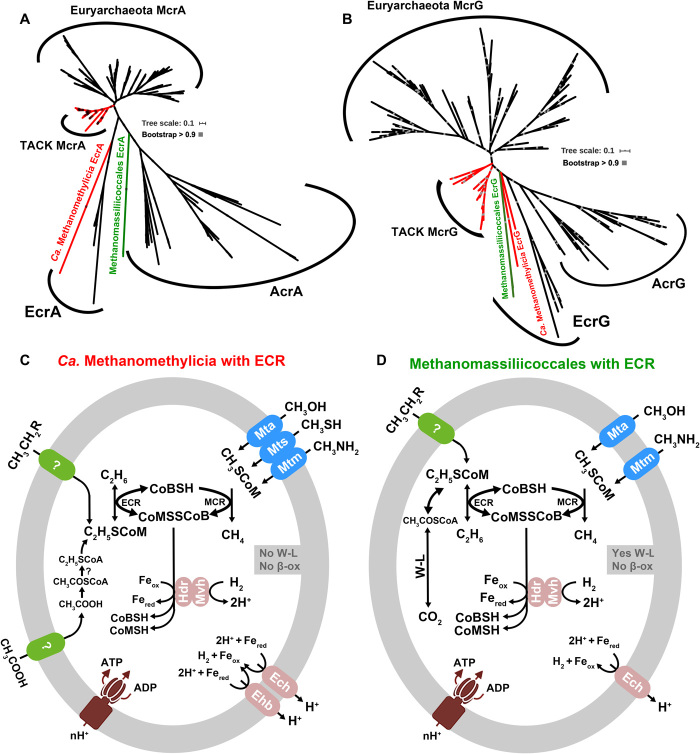
Fig. 2. Phylogeny of MCR/ECR/ACR subunits and metabolic models of studied organisms.
(A) Phylogenetic analyses of McrA/EcrA/AcrA sequences. (B) Phylogenetic analyses of McrG/EcrG/AcrG sequences. McrAG/EcrAG/AcrAG phylogenetic alignments are based on MAFFT and then filtered with trimAl, and the trees were built by the IQ-Tree method with model LG+C60+F+G using SH approximate likelihood ratio test implemented with 1000 bootstrap replicates. (C) Metabolic model of MCR- and ECR-containing Ca. Methanomethylicia, one MAG with the completeness of 100% and contamination of 0.93%. (D) Metabolic model of MCR- and ECR-containing Methanomassiliicoccales, with the completeness of 83.24% and contamination of 6.45%. They contain both MCR and ECR, as well as methyltransferases and hydrogenases, and therefore have the potential to use methylated compounds to produce methane. MAGs from Ca. Methanomethylicia lack Wood-Ljungdahl (W-L) and beta-oxidation (β-ox) pathways, while the MAG from Methanomassiliicoccales has a bacterial-type Wood-Ljungdahl pathway but lacks a beta-oxidation pathway. Their ECR may enable them to gain energy from the reduction of acetate or ethyl compounds to produce ethane or oxidize ethane or propane, etc., using unknown pathways.