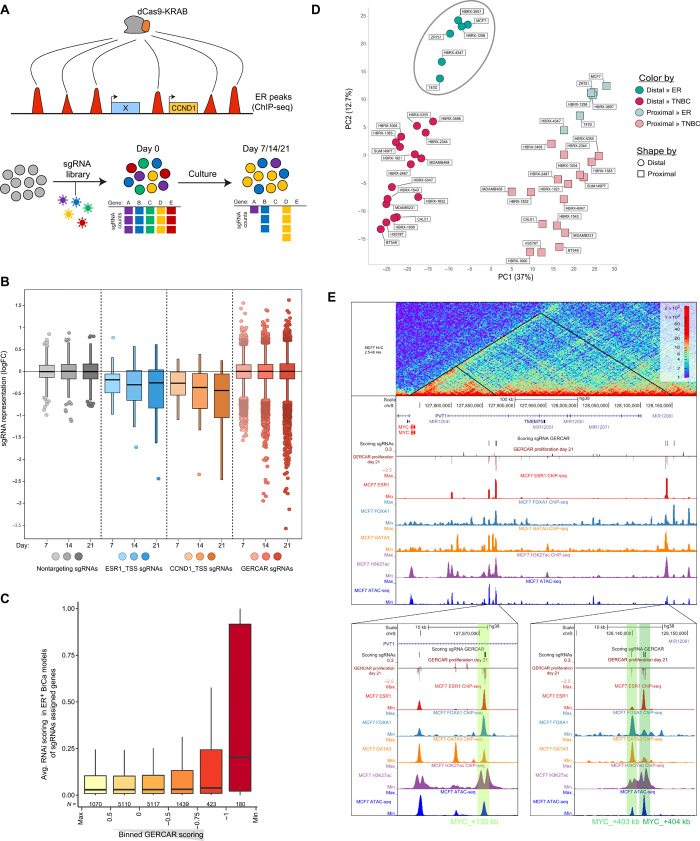
Fig. 2. Genome-scale CRISPRi screen identifies essential ERBS in breast cancer cells.
(A) Schematic representation of the GERCAR screen. (B) Box plot showing the representation of the GERCAR library at days 7, 14, and 21. CCND1_TSS and ESR1_TSS sgRNAs are positive controls. Nontargeting sgRNAs are negative controls. Data represent median logFC of two experiments. (C) Average RNAi score (DEPMAP-combined RNAi) of genes assigned to the closest sgRNA binned by GERCAR scoring (logFC at T = 21 days). (D) PCA of H3K27ac signal (log10) at the 2000 (union of 1000 distal and 1000 proximal) most variable sgRNA targeting regions in cell lines and primary tumor xenografts (HBRX, human breast xenograft). Distal, >5 kb from annotated TSS; proximal, <5 kb from annotated TSS. (E) Snapshot of the MYC locus showing Hi-C data (top track); the GERCAR screen results; ChIP-seq signal of ER, FOXA1, GATA3, and H3K27Ac; and ATAC-seq in MCF7 cells.