Figure 1. Sequence alignment and CA structure.
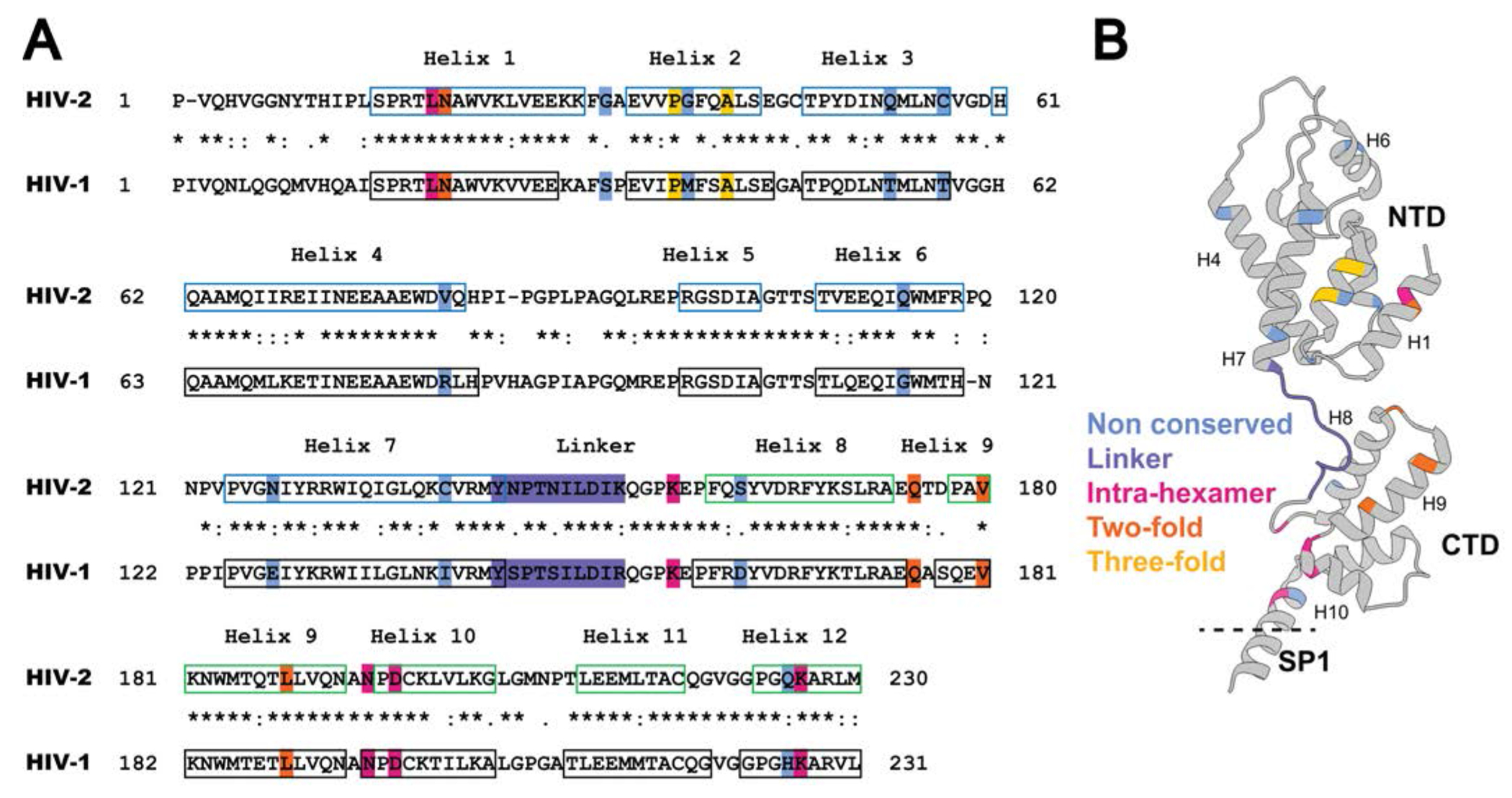
(A) Alignment of the HIV-1 and HIV-2 CA sequences. Amino acid sequences in CA of HIV-1 NL4-3 (GenBank accession no. AF324493; CA 1-231; Gag 133-363) and HIV-2 ROD (GenBank accession no. M15390; CA 1- 230; Gag 136-365) were aligned by using the Clustal Omega Multiple Sequence Alignment [57]. Locations of the HIV-1 immature CA helices in the alignment are based on previous studies [22] (PDB ID: 5L93) and are indicated by black boxes. The HIV-2 immature CANTD (CA 1-144; Gag 136-279) helices were previously described [58] (PDB ID: 2WLV) and are indicated with blue boxes. The CACTD (CA 145-230; Gag 280-365) helices are predicted by the PSIPRED server [59, 60], and are indicated by green boxes. “*” indicates conserved amino acids; “:” indicates amino acid substitution with high amino acid similarity; “.” indicates amino acid substitutions with low similarity. The blue, purple, magenta, orange, and gold-shaded amino acid residues highlight mutagenesis at non-conserved residues, linker, intra-hexamer interface, two-fold interface, and three-fold interface, respectively. (B) Ribbon diagram of the CA monomer structure. PDB ID: 5L93. Shown are the CA amino-terminal domain (NTD), the carboxy-terminal domain (CTD), the spacer peptide 1 (SP1), and the alpha-helices 1, 4, 6, 7, 8, 9, and 10 (i.e., H1, H4, H6, H7, H8, H9, and H10). The non-conserved, linker, intra-hexamer, two-fold and three-fold residues analyzed are indicated on the ribbon structure.
