Figure 2.
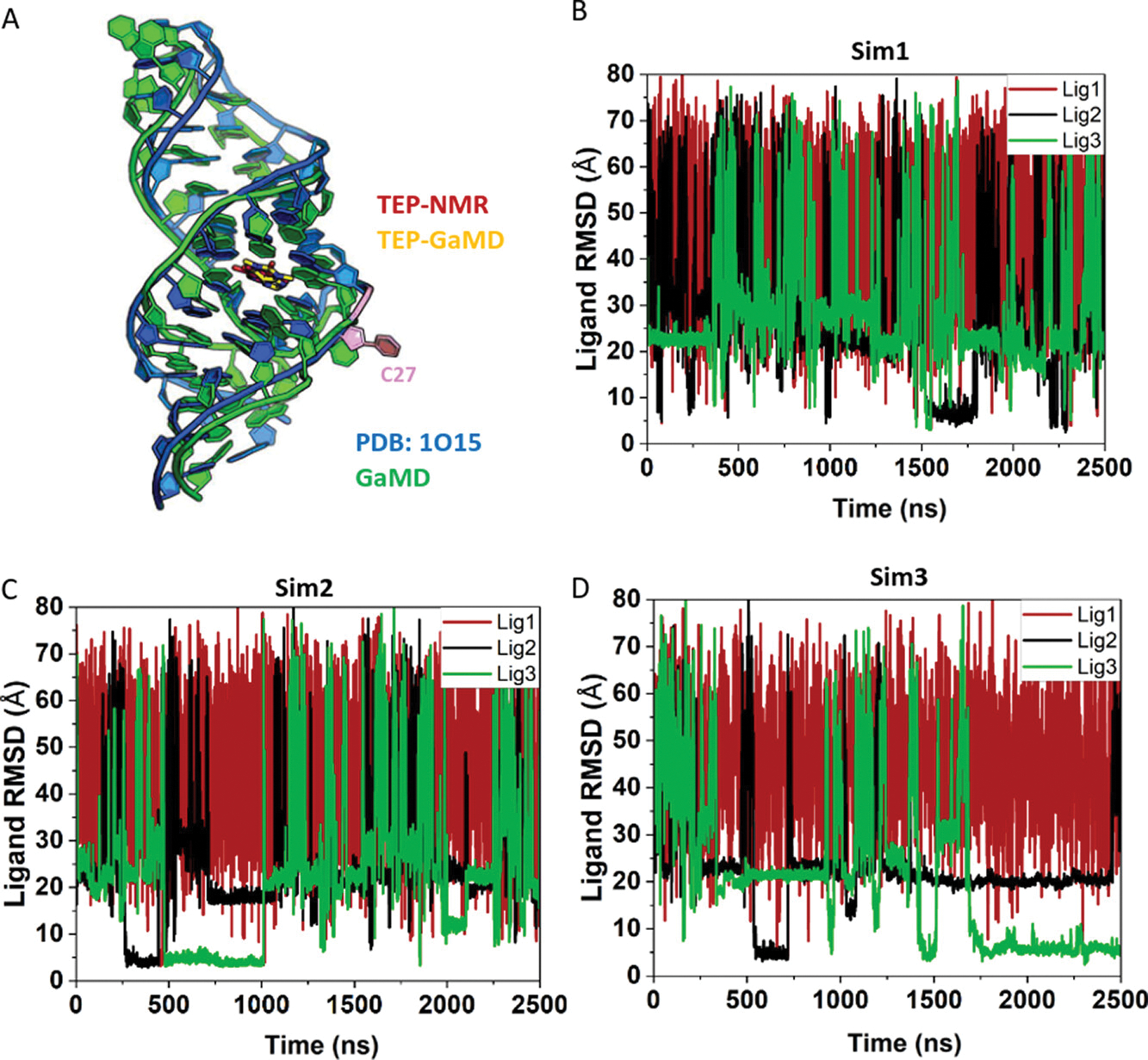
LiGaMD simulations captured repetitive dissociation and binding of theophylline (TEP) to the RNA aptamer: (A) NMR structure (blue, PDB ID: 1O15) and GaMD predicted binding pose of TEP (sticks with C atoms colored in red and yellow, respectively) in the RNA aptamer. A key residue in the binding pocket, C27 is highlighted in light pink. (B–D) Time courses of the ligand heavy atom root-mean-square deviation (RMSD) relative to the NMR structure with the RNA aligned calculated from three independent 2.5 μs LiGaMD simulations of TEP binding to the RNA aptamer.
