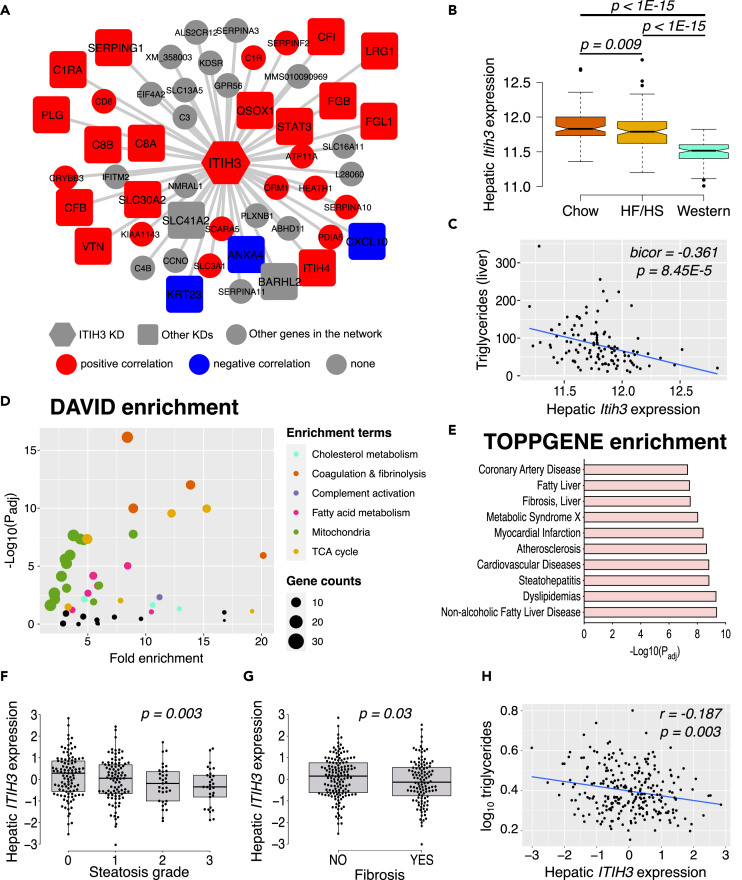
Figure 1.
ITIH3 is negatively associated with NAFLD/NASH in both mice and humans
(A) Overlay of HMDP bicorrelation directionality on ITIH3 key driver (KD) network from our previous study.15 ITIH3 is represented in hexagon shape, other key driver genes are represented in square shapes, and the rest of the network genes are represented in circle shapes. Red represents positive and blue represents negative correlation with liver Itih3 expression from HMDP strains maintained on HF/HS diet (n = 113 HMDP strains).
(B) Liver Itih3 expression from HMDP strains maintained on chow (n = 96 HMDP strains) or HF/HS (n = 113 HMDP strains) or western (n = 102 HMDP strains) diet.
(C) Correlation plot between hepatic Itih3 expression and triglyceride in HMDP strains maintained on HF/HS diet (n = 113 HMDP strains).
(D) DAVID pathway and (E) ToppGene disease enrichment analyses of highly correlated HMDP liver genes (listed in Table S1) with liver Itih3 expression (bicor > |±0.3|; p < 1E-05). Hepatic ITIH3 expression from KOBS cohort (n = 262) grouped by (F) steatosis grade and (G) Fibrosis.
(H) Correlation plot between hepatic ITIH3 expression and triglycerides. Data are presented as median and interquartile range (boxplots). p values were calculated by (A and C) bicor; (B) one-factor ANOVA corrected by post-hoc “Holm-Sidak’s” multiple comparisons test; (F and G) ANCOVA corrected for age, BMI, and sex; (H) partial correlation adjusting for age, BMI, and sex. HMDP, hybrid mouse diversity panel; bicor, biweight midcorrelation; BMI, body mass index.