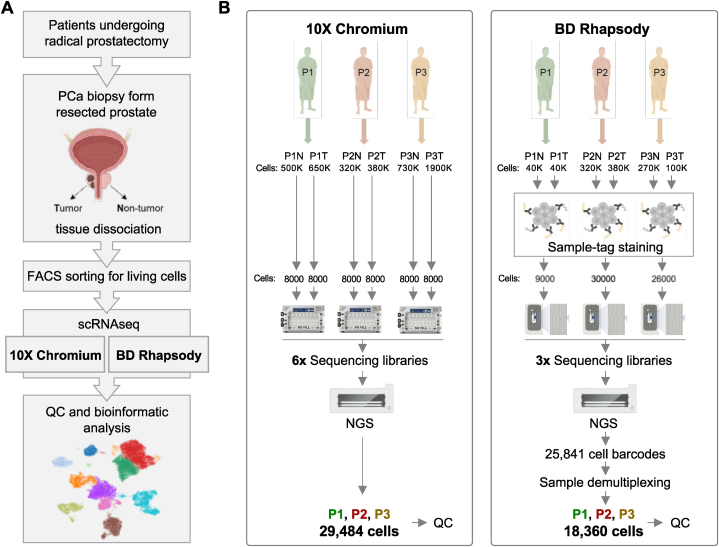
Fig. 1.
Experimental setup. (A) Summary of the analysis workflow. (B) scRNA-seq datasets were generated from freshly isolated benign prostate (n = 3) and PCa (n = 3) tissues using the 10X Chromium and BD Rhapsody platforms, respectively. Cell numbers are shown for each step starting with number of isolated cells from each sample used for the two platforms. In the 10X Chromium dataset 29,484 cells (cells with >100 genes expressed in ≥3 cells) were detected and subjected to quality control processing. In the BD Rhapsody dataset 25,841 cell barcodes were identified and 21,196 cell barcodes with sample-tag information could be recovered during sample demultiplexing. Thereof, 18,360 cells (cells with >100 genes expressed in ≥3 cells) were subjected to quality control processing.