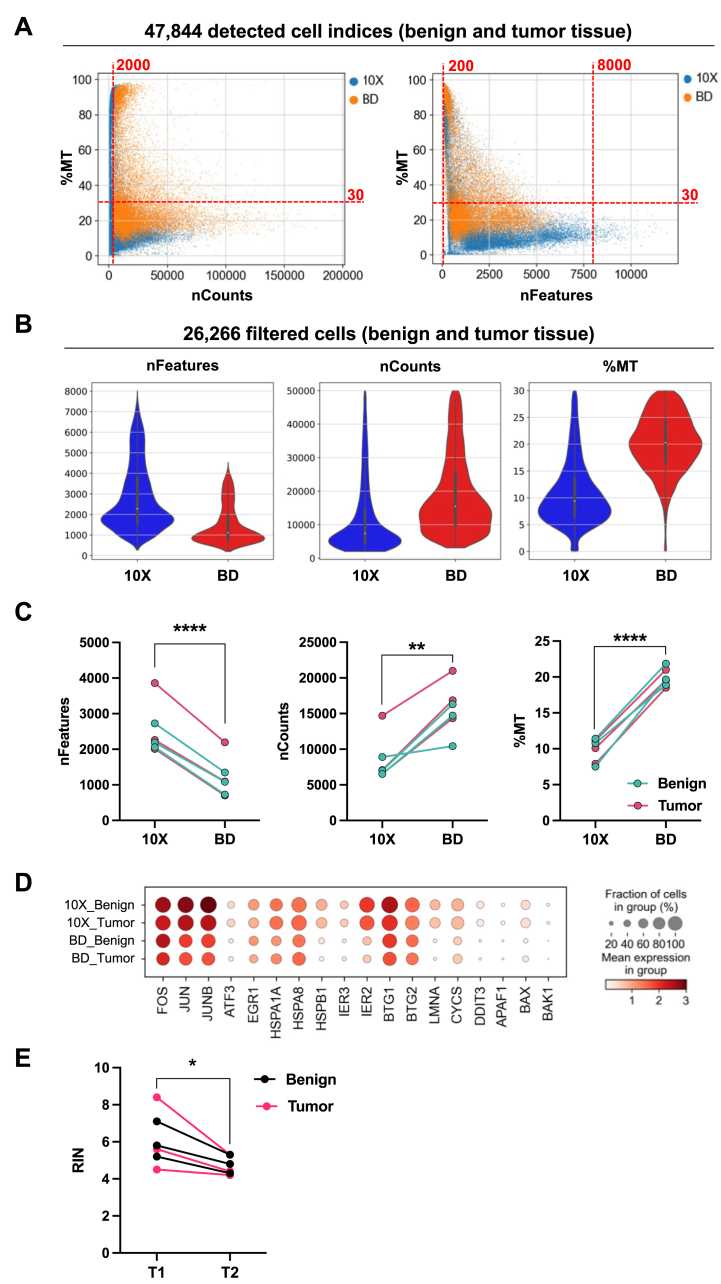
Fig. 2.
QC metrics in datasets generated with 10X Chromium and BD Rhapsody. (A) Correlation of %MT with nCounts and nFeatures quality metrics in data generated with 10X Chromium and BD Rhapsody using cells expressing >100 genes (features). Applied cut-off values to filter for high quality cells are indicated (nCounts >2000, nFeatures >200 and < 8000, %MT < 30%). (B) nFeature, nCount, and %MT quality metrics in filtered cells derived from benign and PCa tumor tissues. (C) nFeature, nCount, and %MT levels in individual samples processed with 10X Chromium and BD Rhapsody (n = 6; benign n = 3, tumor n = 3). Paired t-test, **p ≤ 0.01, ****p ≤ 0.0001. (D) Expression of stress-related transcripts in 10X Chromium and BD Rhapsody data generated from benign prostate and PCa samples. (E) RNA quality (RIN) before (T1) and after (T2) the sample-tag staining procedure in freshly isolated lung cancer (NSCLC) tumor tissues (n = 6). Paired t-test, *p ≤ 0.05.