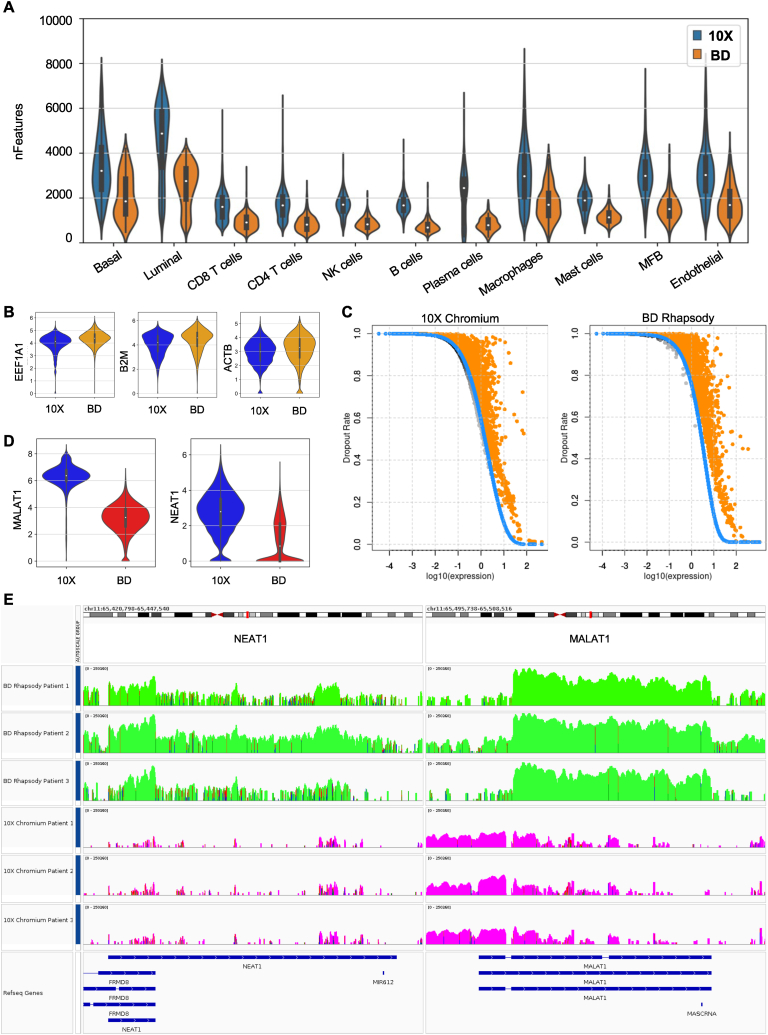
Fig. 4.
Molecule detection efficiency and sequencing library complexity. (A) Number of genes detected per cell in individual cell types depicted by 10X Chromium and BD Rhapsody. (B) Gene expression levels of indicated house-keeping genes in datasets generated with 10X Chromium and BD Rhapsody. (C) Dropout ratios as a function of log10 expression for 10X Chromium (left) and BD Rhapsody (right). Orange dots represent the significant features under the DANB model (at 1% FDR) while gray dots represent the non-significant features. Blue dots represent the expected dropout probabilities as returned from the DANB model. (D) Gene expression levels of MALAT1 and NEAT1 in datasets generated with 10X Chromium and BD Rhapsody. (E) Gene body read coverage of NEAT1 (left) and MALAT1 (right). BD Rhapsody samples are represented with green and 10X Chromium samples with purple colours. Data range is set to 0–250360 (the min - max read output of both platforms for the specific genomic regions) and a log scale is used to visualize the data and to allow comparisons. The blue and red bands indicate mismatches between the reads and the reference genome's nucleotide sequences. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)