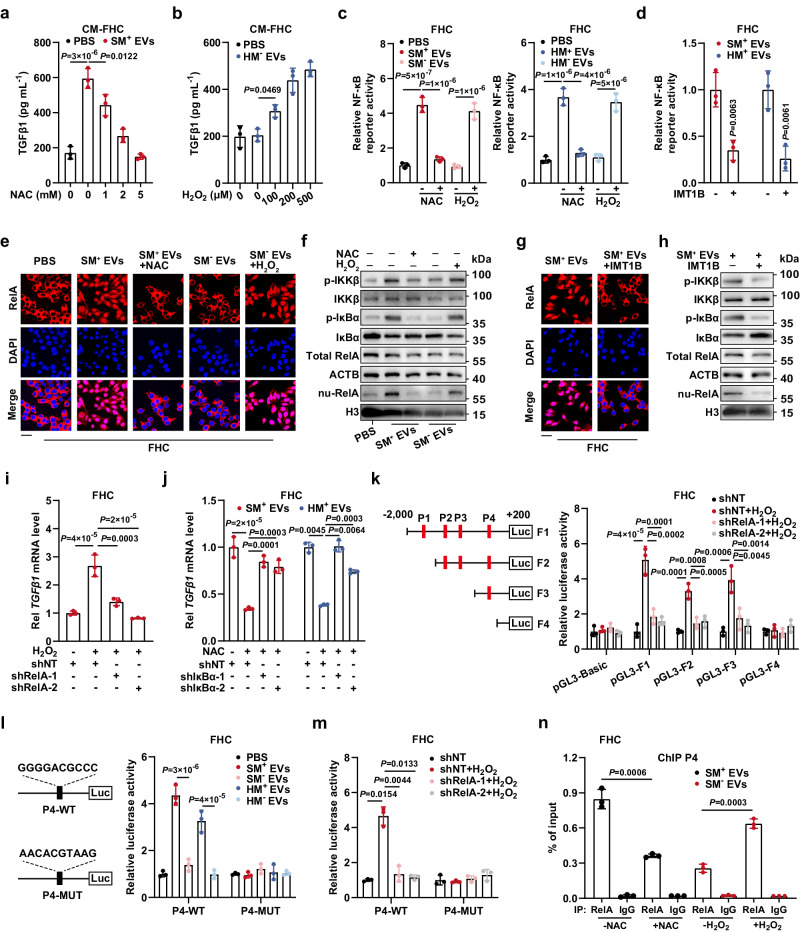
Fig. 7. EV-mtDNA transfer promotes TGFβ1 transcription via ROS-mediated activation of the NF-κB pathway.
a ELISAs were performed to quantify TGFβ1 in CM from FHC. n = 3 independent experiments. b TGFβ1 in CM from FHC in the presence or absence of H2O2 stimulation was quantified. n = 3 independent experiments. c NF-κB reporter activity was measured. Subsets of FHC cells were treated with NAC (5 mM) or H2O2 (500 μM). n = 3 independent experiments. d NF-κB reporter activity in FHC with or without IMT1B treatment (1 μM) was measured. n = 3 independent experiments. The RelA activation was evaluated by (e) immunofluorescence and (f) immunoblotting. Scale bar, 30 μm. The RelA activation was evaluated by (g) immunofluorescence and (h) immunoblotting. Scale bar, 30 μm. i FHC cells expressing shNT or shRelA were stimulated with H2O2. TGFβ1 mRNA levels were then determined. Rel, relative. n = 3 independent experiments. j FHC cells expressing shNT or shIκBα were treated with NAC. Subsequently, TGFβ1 mRNA levels were determined. Rel, relative. n = 3 independent experiments. k The binding site truncation mutants inserted into the pGL3 vector were transfected into FHC cells. The luciferase activity was monitored. n = 3 independent experiments. l The luciferase activity of reporters containing the wild-type or mutated P4 binding site was determined. n = 3 independent experiments. m Luciferase reporter plasmids containing the wild-type or mutated P4 binding site were transfected into FHC cells. Then, luciferase activity was measured. n = 3 independent experiments. n ChIP assays were performed to evaluate RelA enrichment on P4. n = 3 independent experiments. As shown in f and h, the total protein samples derive from the same experiment but different gels for IKKβ, p-IκBα, and another for p-IKKβ, total RelA, ACTB, and IκBα were processed in parallel. The nuclear protein samples for detecting nu-RelA and H3 were processed on the same gel. Data are means ± SD. Two-tailed t test (d, n). One-way ANOVA with Tukey’s (a–c, i–l) or Games-Howell’s (j, m) multiple comparisons test (two-sided). Source data are provided as a Source Data file.